LRP1 gene overexpression in atherosclerotic plaque is associated with increased lipid uptake through the vascular wall. The aim of the study was to analyze whether LRP1 modulates the genetic risk of developing premature cardiovascular disease in familial hypercholesterolemia, using single nucleotide polymorphism association analysis.
MethodsTen polymorphisms of the LRP1 gene (rs715948, rs1799986, rs1800127, rs7968719, rs1800176, rs1800194, rs1800181, rs1140648, rs1800164, and rs35282763) were genotyped in 339 patients (77 with premature cardiovascular disease and 262 without) in the SAFEHEART study.
ResultsThe c.677C>T (rs1799986) polymorphism showed a significant association with premature cardiovascular disease after adjusting by sex, age, body mass index, and the effect of the low-density lipoprotein receptor mutation in the dominant model (CT+TT vs CC: odds ratio=1.94; 95% confidence interval, 1.08-3.48; P=.029). Similar results were observed after increasing the sample to 648 subjects (133 with premature cardiovascular disease vs 515 without [odds ratio=1.83; 95% confidence interval, 1.16-2.88; P=.011]).
ConclusionsThe c.677C>T polymorphism is associated with increased rates of premature cardiovascular disease in familial hypercholesterolemia. Although we were unable to show that this polymorphism was involved in the alteration of normal mRNA splicing patterns, the possibility that it is in strong linkage disequilibrium with another functional polymorphism cannot be ruled out and would explain the cause-effect relationship with cardiovascular disease risk in this population. Further studies are needed to replicate the results and to localize the putative genetic variants associated with this polymorphism.
Keywords
Familial hypercholesterolemia (FH) is characterized by high levels of low-density lipoprotein cholesterol in plasma. It leads to high cholesterol deposits in tissue, accelerated atherosclerosis, and an increased risk of premature cardiovascular disease (PCVD).1 The most common genetic anomaly consists of mutations in the low-density lipoprotein receptor (LDLR) gene; the population frequency of heterozygous familial hypercholesterolemia (hFH) is approximately 1/500. Although genetically the disease is caused by mutations in the LDLR gene, the clinical phenotype of FH can vary regardless of the mutation and this variability is assumed to be due to both environmental and genetic factors.2, 3
Discovering variations in other genes which may modify susceptibility to cardiovascular events is of great interest because of their possible use as predictors of disease.4 The genes involved in cholesterol uptake and homeostasis are good candidates for this type of study. The low-density lipoprotein receptor-related protein-1 (LRP1) is a multiligand transmembrane receptor belonging to the LDLR family which binds low-density lipoproteins and facilitates the removal of a wide variety of ligands involved in fibrinolysis, atherogenesis, and thrombogenesis.5, 6, 7 Overexpression of LRP1 in atherosclerotic plaque has been demonstrated in animal models and humans.8, 9 Several studies have shown a relationship between alterations in LRP1 expression and coronary heart disease.8, 10 Overexpression has also been described in the protein and mRNA of patients with homozygous FH.11 Overall, published results indicate that LRP1 may be a key receptor in the pathogenesis of atherosclerosis.
The aim of this study was to analyze whether there are LRP1 gene polymorphisms which are associated with the occurrence of PCVD in FH.
Methods PopulationThe study initially included 339 cases of unrelated heterozygous FH patients from the SAFEHEART study12, 13 (“Methods” in the supplementary material). Patients were assigned a diagnosis of cardiovascular disease (CVD) if there was a history of myocardial infarction, coronary bypass or angioplasty, angina pectoris with angiographically diagnosed coronary atherosclerosis (stenosis> 50%), stroke, or a peripheral vascular event. Cardiovascular disease was considered PCVD if the event occurred before 55 years of age in men and before 65 years of age in women.14 For each patient, data were collected on blood lipid levels, hypertension, diabetes mellitus, and smoking; anthropometric data were also recorded. The characteristics of the study population are shown in Table 1; LDLR mutations were identified in all patients. Details of the mutations found and their classification based on residual LDLR activity (null, deficient, or indeterminate) can be found in “Methods” and Table 1 in the supplementary material.
Table 1. Characteristics of Study Patients With Heterozygous Familial Hypercholesterolemia, Grouped by Premature Cardiovascular Disease
| No PCVD (n=262) | PCVD (n=77) | P | |
| Men | 111 (42.4) | 47 (35.9) | .004 |
| Women | 151 (57.6) | 30 (41.1) | |
| Age, years | 45.2±15.3 | 53.19±11.3 | <.0001 |
| BMI | 26.3±4.8 | 28.01±5.0 | .014 |
| Hypertension | 42 (16.1) | 20 (26) | .06 |
| Smokers | 115 (43.8) | 42 (54.5) | .067 |
| Diabetes | 11 (4) | 3 (4) | .98 |
| Total cholesterol, mg/dL | 287.1±62.1 | 281.1±70.2 | .512 |
| LDL cholesterol, mg/dL | 215.3±58.7 | 209±67.7 | .925 |
| HDL cholesterol, mg/dL | 52.2±13 | 51.3±17.4 | .24 |
| Triglycerides, mg/dL | 92.5 (38-320) | 94.5 (44-258) | .589 |
| ApoA, mg/dL | 141.1±35.2 | 138.8±32.1 | .204 |
| ApoB, mg/dL | 166.4±43.9 | 159.4±41 | .256 |
| Effect of the mutation on LDLR activity | .527 | ||
| Null allele | 161 (61.6) | 51 (66.2) | |
| Low activity | 75 (28.5) | 22 (28.6) | |
| Undetermined activity | 26 (9.9) | 4 (5.2) |
ApoA, apolipoprotein A; ApoB, apolipoprotein B; BMI, body mass index; HDL, high density lipoprotein; LDL, low density lipoprotein; LDLR, low-density lipoprotein receptor; PCVD, premature cardiovascular disease.
Results are shown as mean±standard deviation, no. (%) or medians (range).
Statistical significance was set at P<.05.
We first searched for polymorphisms in the LRP1 promoter through single-strand conformation polymorphisms and sequencing in a sample of 86 patients with hFH. We then analyzed the genetic association of 10 polymorphisms with PCVD; DNA was extracted from peripheral blood using the Wizard® Genomic DNA Purification kit (Promega).
Screening of Polymorphisms in the LRP1 PromoterSee “Methods” and Table 2 in the supplementary material.
Genotypic Association StudyWe selected 10 LRP1 single nucleotide polymorphisms (SNPs) together with the polymorphism found in the promoter, on the basis of their frequency, bibliographic references, and functionality (Table 2 and Figure 1). The polymorphisms were genotyped using a TaqMan®-based polymerase chain reaction assay and allelic discrimination with an Applied Biosystem 7900 apparatus.
Table 2. Characteristics of the Polymorphisms Analyzed
| Polymorphism analyzed | Genomic variants | SNP ID dbSNP | Alleles 1>2 a | Genomic localization | Functional effect | Published MAF b | MAF in this study |
| 1 | c.1-25C>G | rs35282763 | C>G | Promoter | New Sp1 sequence | 0.08 | 0.08 |
| 2 | IVS2+617C>T | rs715948 | C>T | Intron 2 | — | 0.32 | 0.31 |
| 3 | c.677C>T | rs1799986 | C>T | Exon 3 | Asp100Asp | 0.14-0.15 | 0.14 |
| 4 | c.1116C>T | rs1800127 | C>T | Exon 6 | Ala217Val | 0.03 | 0.03 |
| 5 | IVS6+245C>G | rs7968719 | G>C | Intron 6 | — | 0.49 | 0.47 |
| 6 | IVS19+144C>T | rs1800176 | C>T | Intron 19 | — | 0.30 | 0.30 |
| 7 | c.4012C>T | rs1800194 | C>T | Exon 22 | Cys1182Cys | 0.31 | 0.30 |
| 8 | IVS38+53C>T | rs1800181 | C>T | Intron 38 | - | 0.30 | 0.30 |
| 9 | c.10249G>A | rs1140648 | G>A | Exon 61 | Thr3261Thr | 0.28 | 0.30 |
| 10 | IVS83+139a>G | rs1800164 | G>A | Intron 83 | — | 0.28 | 0.35 |
dbSNP: single nucleotide polymorphism database; MAF: minor allele frequency; SNP ID: single nucleotide polymorphism identification.
a Allele 1 occurs most frequently.
b Minor allele frequency in the HapMap CEU or CEPH panel, found in the literature. 8,15,16
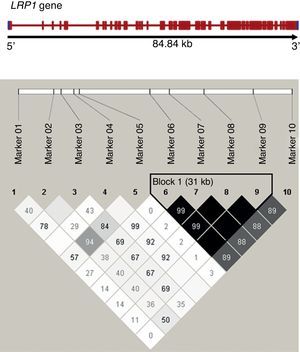
Figure 1. Linkage disequilibrium analysis between analyzed LRP1 single nucleotide polymorphisms. Haplotype blocks defined using Gabriel's method (Haploview 4.0), with the linkage disequilibrium between pairs of markers shown as r2 (in black r2>0.8). The frame to the right of the diagram shows a block with strong linkage formed by single nucleotide polymorphisms 6 to 9 of the study.
Functional Analysis of the c.677C>T Polymorphism (rs1799986)The possible functional effect of the polymorphism on mRNA processing was analyzed using bioinformatic support and in vitro minigene analysis. In this test, the exon of interest is cloned between two synthetic exons for subsequent analysis of RNA processing (“Methods” in the supplementary material).
Statistical AnalysisClinical data were compared between groups using the Student t test for continuous variables, χ2 for categorical variables, and nonparametric tests for non-normally distributed variables (SPSS v.14.0). Allele frequencies were calculated from the genotypes of the subjects. For each polymorphism, we used χ2 to test whether distribution met the Hardy-Weinberg equilibrium, and we used multivariate logistic regression to analyze the association between LRP1 gene polymorphisms and PCVD in hFH, as well as to analyze interaction with covariates.17, 18 We calculated the odds ratio (OR) and 95% confidence intervals (95%CI) for each genotype compared to the homozygote for the most frequent or reference allele, and we estimated significance using a threshold of P<.05 for a general model of inheritance (3 separate genotypes) and for the dominant model.
The prior statistical power of the sample (77 cases and 262 controls) was estimated at 80% for a disease prevalence of 20% and an effect of the polymorphism on the risk of a cardiovascular event >2, and >1.7 for allelic frequencies of 0.14 and 0.30, respectively (Quanto v 1.2). We included markers with lower allele frequencies because of their functionality (promoter polymorphism, SNP1, and the SNP4 amino acid change mutation).
All analyses were adjusted for age, sex, and other CVD risk factors in the study population (Table 1). Haplotype frequencies were estimated by linkage disequilibrium (LD) blocks defined by the Gabriel method, and the values of LD and r2 between SNP pairs were calculated using Haploview v. 4.2 (Figure 1).
Results Characteristics of the Study Population by Presence or Absence of Premature Cardiovascular DiseaseThe characteristics of the study population according to the presence or absence of PCVD are shown in Table 1. At the time of enrollment in the study, 86.5% of patients with hFH were receiving lipid-lowering therapy. Coronary artery disease or ischemic heart disease accounted for 93.4% of the events. There was a higher predisposition to PCVD in men (P=.004), in older patients (P<.001), and in those with a higher body mass index (BMI) (P=.01). There were no significant differences in the proportion of smokers in each group at the time of inclusion in the study.
Some sex-based differences were observed (data not shown in Table 1): BMI was 28.4 (6.6) in women with PCVD vs 26.7 (5.7) in women without PCVD (P=.137) and 27.7 (3.7) in men with PCVD compared to 25.9 (3.1) in men without PCVD (P=.003). There were no significant differences in the frequency of hypertension and smoking between both groups, although hypertension was more prevalent in women (40% in PCVD vs 18.2% in non-PCVD, P=.014) than men (16.3% vs 13.2%, P=.612) and smoking was more prevalent in men (82.6% in PCVD vs 58.7% in non-PCVD, P=.005) than women (16.7% vs 33.7%, P=.085). While the percentage of individuals with PCVD was higher among patients with hFH and null-allele or more severe mutations in LDLR, the differences were not significant in this sample.
Screening of Polymorphisms in the LRP1 PromoterWe detected a polymorphism in the promoter (Fig. 1A in supplementary material). According to the in silico analysis, this polymorphism (c.1-25C>G [rs35282763]) creates a new binding site for the Sp1 transcription factor (Fig. 1B, supplementary material).
Association of Polymorphisms and Haplotypes With Premature Cardiovascular Disease in Heterozygous Familial HypercholesterolemiaThe distribution of genotypes conformed to the Hardy-Weinberg equilibrium in all analyzed polymorphisms (P>.2). Allele frequencies observed in the population did not differ significantly from previously published data (Table 2). In addition, markers 2, 3, 5, and 6 are tag SNPs according to the HapMap3 Genome Browser, release #2.
The results of the association with PCVD after adjusting for age, sex, BMI, and the effect of LDLR mutation are shown in Table 3. Only SNP3 (rs1799986) showed a significant association with PCVD in the dominant model (CT + TT vs CC, OR=1.94; 95% Cl, 1.08-3.48; P=.029). We obtained similar results after increasing the sample to 648 hFH patients (133 with PCVD, and 515 without PCVD) (Table 3 in supplementary material) and reanalyzing the data (OR=1.83; 95%CI, 1.16-2.88; P=.011).
Table 3. Association of Polymorphisms and Premature Cardiovascular Disease in Heterozygous Familial Hypercholesterolemia
| SNP | SNP ID | HFh (n=339) | Genotypic frequencies | POR (95%CI) | ||
| 1 | rs35282763 | CC | CG | GG | .9 | |
| No PCVD | 0.84 | 0.15 | 0.04 | |||
| PCVD | 0.88 | 0.11 | 0.01 | |||
| 2 | rs715948 | CC | CT | TT | .86 | |
| No PCVD | 0.5 | 0.38 | 0.13 | |||
| PCVD | 0.51 | 0.39 | 0.1 | |||
| 3 | rs1799986 | CC | CT | TT | .029 | |
| No PCVD | 0.77 | 0.2 | 0.02 | 1.94 (1.08-3.48) | ||
| PCVD | 0.64 | 0.36 | 0 | CT+TT vs CC | ||
| 3 * | rs1799986 | CC | CT | TT | .011 | |
| No PCVD | 0.76 | 0.22 | 0.02 | 1.83 (1.16-2.88) | ||
| PCVD | 0.66 | 0.34 | 0 | CT+TT vs CC | ||
| 4 | rs1800127 | CC | CT | TT | NS | |
| No PCVD | 0.95 | 0.05 | 0 | |||
| PCVD | 1 | 0 | 0 | |||
| 5 | rs7968719 | CC | CG | GG | .79 | |
| No PCVD | 0.29 | 0.47 | 0.24 | |||
| PCVD | 0.33 | 0.39 | 0.28 | |||
| 6 | rs1800176 | CC | CT | TT | .096 | |
| No PCVD | 0.46 | 0.45 | 0.09 | |||
| PCVD | 0.48 | 0.49 | 0.03 | |||
| 7 | rs1800194 | CC | CT | TT | .07 | |
| No PCVD | 0.47 | 0.43 | 0.1 | |||
| PCVD | 0.48 | 0.49 | 0.03 | |||
| 8 | rs1800181 | CC | CT | TT | .07 | |
| No PCVD | 0.47 | 0.43 | 0.1 | |||
| PCVD | 0.48 | 0.49 | 0.03 | |||
| 9 | rs1140648 | CC | CT | TT | .075 | |
| No PCVD | 0.46 | 0.45 | 0.1 | |||
| PCVD | 0.48 | 0.49 | 0.03 | |||
| 10 | rs1800164 | GG | AG | AA | .72 | |
| No PCVD | 0.42 | 0.45 | 0.13 | |||
| PCVD | 0.40 | 0.48 | 0.12 | |||
95%CI, 95% confidence interval; HFh, heterozygous familial hypercholesterolemia; ID, identification; NS, not significant; OR, odds ratio; PCVD, premature cardiovascular disease; SNP, single nucleotide polymorphism.
All values adjusted for age, sex, body mass index and type of low-density lipoprotein receptor mutation; rs1799986 showed a significant P value (dominant model, genotypes CT+TT vs CC).
* Results after increasing the sample to 515 subjects without PCVD and 133 with PCVD.
The allele frequency for rs1799986 polymorphism in the population with HF was similar to that of the Spanish control population (P=.76) and other Caucasian populations (see “Methods” and Table 4 in supplementary material).
We analyzed the interaction of the rs1799986 polymorphism with the type of LDLR mutation (null allele, deficient activity, or indeterminate), and found a higher percentage of null-allele mutations in individuals with the polymorphism and PCVD compared to patients without PCVD. However, the results are not conclusive due to the sample size (Table 5 in supplementary material).
Using Haploview, we examined the existence of blocks of polymorphisms with a strong linkage disequilibrium. We observed only one block, formed by SNP 6 to 9 according to Gabriel's method (Figure), but no haplotype associated with the disease (Table 4).
Table 4. Association of Haplotypes With Premature Cardiovascular Disease, After Adjusting for Risk factors (n=339)
| Frequency of haplotypes | OR (95%CI) for the most frequent | P | |
| CCCA | 0.692 (0.727-0.682) | 1 | |
| TTTG | 0.298 (0.306-0.293) | 0.84 (0.54-1.31) | .44 |
| General association for haplotypes | .15 |
95%CI, 95% confidence interval; OR, odds ratio.
Frequency of haplotypes in the block with strong linkage formed by single nucleotide polymorphisms 6 to 9, estimated using the maximum expectation algorithm; total frequency (premature cardiovascular disease/no premature cardiovascular disease).
As pointed out in the “Statistical Analysis” section, the initial study sample size only allowed us to detect associations with an OR>2, with 80% power for allele frequencies of 0.14, such as the one with SNP3. Due to continued enrollment of new patients into the study cohort, it was possible to expand the sample size of individuals with hFH to a total of 133 with PCVD and 515 without PCVD (Table 3 in supplementary material). This allowed us to detect a statistically significant OR of 1.83 with 80% statistical power for this allele frequency (Quanto v1.2.0).
The results suggest that the presence of the T allele in polymorphism c.677C>T (rs1799986) is associated with an increased risk of PCVD in hFH.
Functional Analysis of the Polymorphism c.677C>T (rs179998)Functional studies ruled out the possibility that this polymorphism may alter the normal pattern of mRNA processing (see “Results” and Fig. 2 in the supplementary material).
DiscussionPrevious studies have cited the association of LRP1 polymorphisms with coronary artery disease in Caucasian populations,8, 19, 20 with coronary artery disease and longevity,21 and with coronary thrombosis,22, 23 although other studies have presented contradictory or less significant results for such an association in relation to dyslipidemia or Alzheimer disease15 and thrombosis.24 This and other literature on the involvement of LRP1 polymorphisms in complex diseases is included in the summary by Gläser et al.16 Recently, Peters et al.,25 in a comprehensive study of candidate genes involved in the efficacy of cholesterol-lowering statins found that a polymorphism in intron 2 of LRP1 (rs715948, SNP 2 of this study) was associated with a reduction in myocardial infarction. However, this finding was not replicated in our study of hFH using a broader CVD phenotype than the one described above.
There have been conflicting results regarding the involvement of the c.677C>T polymorphism with CVD in Caucasian populations. Pocathikorn et al.,20 in a study of 600 individuals with coronary artery disease and 700 controls, found a significantly lower frequency of the TT genotype of the c.677C>T polymorphism in individuals with coronary heart disease compared to controls. On the other hand, Benes et al.,21 in a study with 654 individuals with coronary artery disease and 525 controls, found that the T allele increased the risk of coronary heart disease in subjects with the 5G/5G plasminogen activator inhibitor-1 genotype.
Although in this study we attempted to analyze polymorphisms throughout the gene, its size of about 85kb meant we would require a larger number of SNPs to cover maximum variability in the region, particularly at the 5’ extreme, where the analysis of region chr12: 5508548..55893389 with Haploview (HapMap3 Genome Browser release #2) indicated a lower density of polymorphisms with LD (results not shown).
LimitationsAmong the limitations of the study, in addition to those derived from the sample size and power of the trial, are those deriving from the number of polymorphisms analyzed and the probability of finding an association by chance. The results should therefore be confirmed through replication in other populations before moving on to search for other polymorphisms in LD with the genetic variant associated with increased risk of PCVD.
The study calculated cardiovascular events that occurred prior to inclusion in the study using age-based guidelines established for PCVD; however, since this was a retrospective cohort, there may have been some selection bias, as some fatal cases may not have been included. An important limitation of the study design is that the phenotype analyzed was based on the end point, ie, the cardiovascular event, so no data were available on the degree of atherosclerotic disease prior to the event, such as number of vessels affected; this would require another type of design and clinical analysis which were not considered in all of the present samples.
As described in the literature,26 the additive effect of several genetic variants may help to better define the risk of ischemic heart disease and, by extension, CVD. It would therefore be interesting to analyze the functional variants of LRP1 together with other genes associated with disease and/or involved in similar processes.
ConclusionsIt appears that the T allele of the c.677C>T (rs179998) polymorphism increases the risk of PCVD in HFh. As the functionality of the polymorphism has not been demonstrated, this will presumably be found in LD with another genetic variant that increases the risk of CVD. Further studies are needed to replicate the results of the present study in other populations and then to determine the polymorphism(s) having a strong LD with rs179998 and an association with cardiovascular disease.
FundingThis study was supported in part by funding from SAF2010-16549, CNIC-08-2008, and CIBERobn CB06/03.
Conflicts of interestNone declared.
Acknowledgements
We would like to acknowledge Nuria Sala, Xavier Muñoz (ICO-IDIBELL), and Mónica Gratacós (CRG) for their contribution to scientific discussion of the paper, Montse Gómez for her technical assistance, and the Foundation for Familial Hypercholesterolemia for their support for the study.
Supplementary MaterialSupplementary material associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.rec.2012.03.012.
Appendix A. Supplementary dataReceived 6 October 2011
Accepted 7 March 2012
Corresponding author: Centro de Investigación Cardiovascular, CSIC-ICCC, Hospital de la Santa Creu i Sant Pau, Sant Antoni Maria Claret 167, 08025 Barcelona, Spain. lbadimon@csic-iccc.org