Calcifying nanoparticles, also known as “nanobacteria,” are very small bacteria-like structures (0.1-0.5μm) with the ability to facilitate the precipitation and growth of calcium phosphate in pathological conditions and have been associated with aortic valve calcification. The status of nanobacteria is controversial; some have proposed that they are a new class of living organism while others describe calcifying nanoparticles as mineralo-fetuin complexes. The objective of the present study is to elucidate if calcifying nanoparticles are living entities, based on whether or not they have metabolic activity, a characteristic of life, irrespective of their composition.
MethodsCalcifying nanoparticles were grown from 6 different valves randomly chosen among 84 consecutively explanted aortic valves, as described in the literature. The 1H-NMR spectra were acquired from calcifying nanoparticles culture media to assess metabolic changes and the presence of 16sRNA in the culture media was investigated by real-time polymerase chain reaction.
ResultsAfter 6 weeks in culture, calcifying nanoparticles could be seen clearly attached to the surface of culture flasks. All samples were negative for 16sRNA, discarding the presence of known bacteria. 1H-NMR spectra showed no difference between calcifying nanoparticles and 6-week-old sterile culture media maintained under the same conditions.
ConclusionsOur results show that calcifying nanoparticles cannot be considered as living organisms.
Keywords
Degenerative aortic valve disease, a progressive disease characterized by dystrophic calcification of the valve cusps and narrowing of the aortic valve orifice,1 is the most common valvulopathy and an important cause of mortality and morbididy. There is growing evidence that the degree of valve calcification correlates with more rapid disease progression and with worse clinical outcomes.2 Valvular calcium deposits contain both calcium and phosphate as hydroxyapatite, the form of calcium-phosphate mineral present in both calcified arterial tissue and bone.3
The term “nanobacteria” is a neologism introduced by Dr. Kajander as the name for very small bacteria-like organisms. The extremely small size (0.1-0.5μm) and the ability to facilitate the precipitation and growth of calcium phosphate in physiological conditions are the principal characteristics of these particles, also known as calcifying nanoparticles (CNP).4 This singularity promoted the idea that nanobacteria could be related with diseases by being linked with calcium deposits. The presence of CNP has been described in a great variety of diseases, including aortic stenosis5 and arterial calcification.6 Intravenous injection of CNP in animals induced accelerated biocrystallization of calcium oxalate monohydrate.7, 8
The status of CNP is still controversial, and has spurred one of the biggest controversies in modern microbiology.4, 6, 9, 10 Some researchers have suggested that they are a new class of living organisms because these CNP appear to be self-replicating in culture,11 they are capable of incorporating radiolabeled uridine, a strong immune response against CNPs was documented,12, 13 and CNPs cause cell death in vitro.4, 6, 11, 14 Most studies looking for evidence of life in CNPs have focused on structural elements, searching for nucleic acids or proteins associated to CNPs, so far without conclusive results.15, 16 Recent articles have described CNPs as mineralo-fetuin complexes.10, 12 However, at least in the case of heart valve derived CNPs, the status as living organisms is still under debate.17
Regardless of their nature and chemical composition, living organisms “preserve their internal order by taking from their surroundings free energy, in the form of nutrients or sunlight, and returning to their surroundings an equal amount of energy as heat and entropy.”18 In other words, the presence of metabolic activity could, on its own, provide evidence of the existence of life. Searching for new life forms has become especially relevant since the advent of space exploration because a main goal of such programs has been to find hallmarks of metabolic activity. For example, during the early seventies the search for life took on renewed interest, mainly during the Viking program of Mars exploration that included experiments designed to search for life based on metabolic activity in the soil of Mars.19
In the case of cultured CNPs, the source of energy must be chemical, from nutrients present in the culture media. In other words, they must possess some sort of metabolic activity. At present, the only reference about CNPs metabolism is that they are “of limited metabolic activity.”20 Metabolomics, being a hypothesis-free approach, is ideal for such an study because is able to detect changes in many metabolites without any prior knowledge of which metabolites could be affected.
The objective of the present work was to investigate the potential biological origin of CNP by analyzing 1H-NMR metabolic profiles of CNP culture media.
Methods Aortic ValvesAortic valves were obtained from 84 patients undergoing valve replacement because of symptomatic aortic valve disease. Valves were obtained consecutively between January 2004 and July 2005. Exclusion criteria were congenital aortic stenosis and bicuspid aortic valve. Six aortic valves with positive culture were randomly selected from the total group for the subsequent nuclear magnetic resonance (NMR) spectroscopy analysis. The study protocol was approved by the Ethics Committee and by the Hospital's Research Committee. No patient initially assessed for study inclusion refused to enter the study.
Echocardiographic assessment was carried out with a Vivid 3 (General Electric, Massachusetts, United States) equipped with a 1.5 to 3MHz transduce, using standardized imaging techniques. The peak velocity across the valve was measured with continous-wave Doppler. Aortic valve area was calculated by the continuity equation. The degree of aortic valve calcification was also assessed and scored as mild (no calcification or small isolated calcified spots) or severe (multiplate larger spots or extensive thickening and calcification of all the aortic valve cusps).21
Culture of Nanosized ParticlesSix aortic valves were randomly selected and cultured as described previously.5 Briefly, the valves were divided into two parts, one of which was frozen at −80°C for future analysis and the other was crushed in a sterile glass mortar. Primary cultures were established by demineralization of the valves by adding 1M HCl, which was subsequently neutralized with 1M NaOH. The resultant was filtered with a 0.22μm pore filter and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with gamma-irradiated fetal bovine serum at 37°C, under an atmosphere with 5% to 10% CO2. Once the culture was established, subsequent passes were obtained in fetal calf serum (FCS)-free DMEM. All culture media used was at least 5 passes old. After 6–8 weeks of culture incubation, fresh medium was used for NMR spectroscopy and frozen in sterile conditions for later polymerase chain reaction (PCR) analysis.
Uncalcified valves do not give rise to CNP growth. As control we used sterile media kept in the same conditions for the same time as CNP cultures.
SpectroscopyMedia from CNP cultures were obtained in sterile conditions and diluted in 1:1 in deuterium oxide (D2O) prior to NMR spectroscopy.
The NMR spectrum of the culture was obtained on a 400MHz vertical bore magnet equipped with a 5-mm probe interfaced to a Bruker Avance spectrometer (Bruker, Madrid, Spain). One-dimensional pulse-and-acquire sequences were used, preceded by a 3-9-19 WATERGATE sequence to reduce the water signal; each spectrum consisted of the accumulation of 64 scans and lasted for approximately 6min. In some samples 2-dimensional 1H-1H correlation spectroscopy and 1H-13C heteronuclear single quantum coherence spectra were acquired for peak assignation purposes. All spectra were acquired at 30°C.
Pattern RecognitionFor pattern recognition, 1-dinensional NMR spectra were digitized in 1000 bins, the area containing the residual water peak between 4.25 and 5.25ppm was removed, and the resulting spectra were fed into SIMCA-P software (Umetrics, Umeå, Sweden) as described previously.22 Principal component analysis (PCA) was used on the spectra database, allowing a reduction in the number of variables (NMR peaks or metabolites in our case) in orthogonal, thus independent, vectors according to the sources of variation, each vector corresponding to a source of variation in descending order.
PCA, being an unsupervised method, does not involve any input from the observer and samples are grouped solely according to the sources of variation within the dataset; this is the unbiased approach we preferred in order to detect any possible changes in the spectra related to metabolic activity.
16sRNA Polymerase Chain ReactionIn order to determine the presence of bacteria in the CNP, the medium was thawed and submitted in sterile condition to real-time (RT) PCR of the V4 hypervariable region of the bacterial 16S rRNA gene. The V4 regions (290bp) were amplified by using universal primers located in highly conserved sequences of the V4 region: V4 F (5’-GCC AGC AGC CGC GGT AA-3’) and V4R_ 805_19 (5’-GAC TAC CAG GGT ATC TAA T-3’). We selected these primers from among several pairs commonly found in the literature because they matched most of the bacterial sequences deposited in the Ribosomal Database Project.23 Cultured valve and aged DMEM thawed media (negative control) were analysed blindly. To generate a standard curve, calculated amounts of a linearized plasmid, in which the V4 region from a control bacterium had been inserted, were used. Plasmid concentration was measured using a NanoDrop ND-1000 Spectrophotometer (Nucliber), and the number of plasmid copies was calculated from the plasmid's molecular weight. Serial dilutions of the plasmid were amplified ranging from 102 to 107 to be able to extrapolate the bacterial number in each sample. Moreover, known positive and negative controls were determined in parallel in each run. Amplification and detection of DNA by real-time PCR were performed with the 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, California, United States) using optical-grade 96-well plates. The PCR reaction was performed in a total volume of 25μL using the Power SYBR Green PCR Master Mix (Applied Biosystems) containing 100nM of each of the universal forward and reverse primers. The reaction conditions for amplification were 2min at 50°C, 10min at 95°C, and 40 cycles of 15s at 95°C and 1min at 60°C. All the samples were carried out in triplicate and mean values were calculated. Melting curves were performed and inspected after amplification to determine the specificity of the PCR reaction.
Statistical AnalysisCategorical variables are reported as absolute values and percentages. Continuous variables are reported as the mean±standard deviation or median and interquartile range. Normal distribution of quantitative variables was verified with the Kolmogorov-Smirnov test. Qualitative variables were compared with the chi-squared test and Fisher exact test. Continuous variables were compared with Student t test or its equivalent for nonparametric tests, Mann-Whitney U, for variables that were not normally distributed.
All data were entered into a database and analyzed with statistical package PASW (v 17.0.2, Chicago, Illinois). Differences were considered statistically significant when 2-sided nominal unadjusted P-values were<.05.
ResultsThe demographic, clinical, and echocardiographic characteristics and biochemical data of the total population and patients whose valves were included in the study are outlined in Table. All baseline data were similar in both subgroups. Only a trend towards male sex (63% vs 100%; P=.09) was noted in selected valves with positive cultures. Our patients characteristics are similar to those described in previous articles.24, 25
Characteristics of the Total Group and Selected Valves With Positive Calcifying Nanoparticles
| Variables | Total | Selected valves | P |
| Valves, n | 84 | 6 | |
| Age, years | 71 [67-76] | 75 [69-77] | .36 |
| Male sex | 53 (63) | 6 (100) | .09 |
| Systolic blood pressure, mm Hg | 131±2 | 129±7 | .74 |
| Diastolic blood pressure, mm Hg | 65±1 | 66±5 | .85 |
| Smoking habit | 10 (12) | 1 (17) | .57 |
| Hypertension | 52 (62) | 5 (83) | .41 |
| Diabetes | 17 (22) | 1 (17) | 1 |
| Dyslipidemia | 39 (47) | 4 (67) | .42 |
| Coronary angiographic disease | 31 (37) | 1 (17) | .41 |
| Medical treatment | |||
| Anticoagulants | 35 (42) | 2 (33) | 1 |
| Beta-blockers | 30 (36) | 3 (50) | .66 |
| Calcium antagonists | 14 (17) | 0 | .58 |
| ACE inhibitors | 21 (25) | 0 | .33 |
| Statins | 39 (46) | 2 (33) | .69 |
| Diuretics | 44 (52) | 2 (33) | .43 |
| Echocardiographic variables | |||
| Transvalvular peak gradient, mm Hg | 76±3 | 77±8 | .96 |
| Severe calcification grade | 42 (50) | 4 (67) | .360 |
| Aortic valve area, cm2 | 0.73±0.3 | 0.77±0.12 | .748 |
| Analytical variables | |||
| Total cholesterol, mg/dL | 187±4 | 188±13 | .968 |
| HDL-C, mg/dL | 50.1±4 | 59±2.7 | .386 |
| LDL-C, mg/dL | 105.7 ±7.5 | 97±0.5 | .671 |
| Triglyceride, mg/dL | 116±7 | 106±12 | .694 |
| Creatinine, mg/dL | 0.9 [0.8-1.1] | 0.9 [0.8-1.15] | .98 |
| Calcium, mg/dL | 9.7 [9.3-10.6] | 8.9 [8.2-9.6] | .14 |
| Leukocyte count, ×109/L | 6.6 [5.7-7.3] | 6.9 [6.3-8.4] | .36 |
| Fibrinogen, mg/dL | 311±13 | 296±51 | .77 |
ACE, angiotensin-converting enzyme; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol.
Categorical variables are presented as proportions, no. (%). Continuous variables are presented as mean±standard error mean and medians [with the 25th and 75th percentiles].
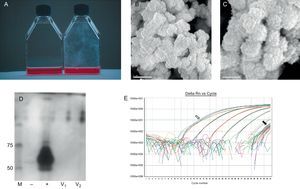
After approximately 6 weeks in culture, CNPs appeared as a film attached to the plastic surface (Figure 1). The culture media remained clear and there was no particular smell or any other characteristic that could be associated with bacterial growth.
Figure 1. A, Photograph of culture flasks with the presence of calcifying nanoparticles (right) and control sterile aged media (left). B and C, Electron micrographs of calcifying nanoparticles (CNP) at different magnification. D, PAGE-SDS gel with lines corresponding to molecular weight markers (M), culture media without and with fetal calf serum as negative and positive controls (−, +), respectively, and culture media from 2 different valves (V1 and V2). E, Real-time polymerase chain reaction plots of CNP culture media with positive controls (open arrow), CNP cultures and negative controls (black arrow), and a standard curve between 107 and 102 copies. All samples and negative controls have CT values above 40.
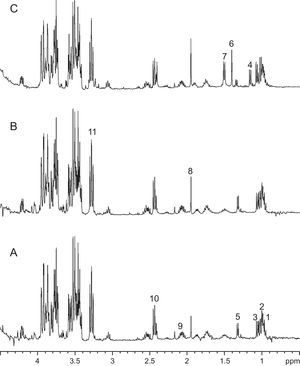
Figure 2 shows NMR spectra of medium obtained after 6 weeks of CNP culture, control medium aged for 6 weeks at 37°C in 5% CO2 atmosphere and medium from a contaminated culture (Figure 2). Spectra from aged media and CNP cultures are similar (Figure 1), with no single metabolite/peak that allows differentiation between them. On the other hand, spectra from contaminated media has peaks at 1.16 and 1.40ppm, tentatively assigned to α-ketoisovaleric and dimethylmalonic acids, respectively, that are not present in the other two.
Figure 2. 1H NMR spectra of aged sterile media (A), culture media after 6 weeks of CNP growth (B) and contaminated media (C). Tentative peak assignation based on chemical shift only is as follows: 1, isoleucine; 2 and 3, valine; 4, α-ketoisovaleric acid; 5, lactate; 6, dimethylmalonic acid; 7, alanine; 8, acetate; 9, glutamate/glutamine; 10, glutamate; 11, glucose.
The other metabolites identified are consistent with those present in the formulation of the culture media. The largest peaks of the spectra correspond to glucose; other metabolites identified include lactate, lysine, glutamine, piruvate, and thyrosine, all of them present in the formulation of DMEM media. We also were able to identify acetate and formate, those compounds appear in aged media, independent of the presence of CNPs, but not in fresh media. These later changes were not related to the presence of known bacterial microorganisms because when culture medium was tested for the presence of 16sRNA bacterial gene the results were negative in all the tested samples (Figure 1E), and consequently not different from the results obtained with the negative control.
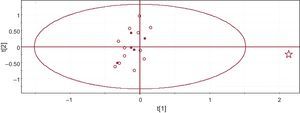
In order to investigate whether inconspicuous differences were present between control and CNPs media, we used the unsupervised pattern recognition PCA approach. Figure 3 shows a PCA score plot, each point corresponding to an individual sample. The only sample falling within the 95% confidence interval is the one corresponding to the contaminated sample (marked as a star in the plot). There was no apparent grouping of samples that allowed us to differentiate between aged media and CNP culture media, further supporting the absence of differences between the 2 sets of media.
Figure 3. Score plot of the principal component analysis of the spectra dataset. Each dot corresponds to an individual culture. Full squares correspond to aged culture media, open circles to media from CNP cultures, and the star to the contaminated media.
DiscussionThis study specifically addressed the question of whether CNPs are a living entity by looking for fingerpints of metabolism in culture media. The results show no metabolic activity that can be directly linked to CNP growth. Accordingly, CNPs should not be considered living organisms according to the definition of live used in the present work.18
As shown in Figure 1, we have been able to grow CNPs with characteristics similar to those described in the literature, both macro- and microscopically,5, 10, 24 This growth has been accomplished without the detectable presence of FCS in the culture media.
There has been controversy around whether CNPs are alive; later results suggest that they are not. Cisar et al.9 concluded that biomineralization attributed to nanobacteria may be initiated by nonliving macromolecules. Later, Raoult et al.11 performed a comprehensive analysis with Nanobacterium sp and concluded that CNPs are “self-propagated mineral protein complexes containing fetuin as the major biological component.” The nonbiological origin of nanobacteria-like particles has also been supported by Martel and Young.24 In addition, unusual crystal growth mechanisms can produce witherite precipitates from barium chloride and silica solutions that closely resemble primitive organisms; thus, evidence for life cannot rest on morphology alone.25 Evidence that CNPs are a live entity is mostly based on their ability to reproduce in culture, but controversy remains. The articles by Wainwright26 and Urbano and Urbano27 are good examples of this controversy.
Our results support recent published data that show CNPs to be a combination of salts and serum proteins11 that can be formed by a non-biological process.28 In these systems the presence of crystals would act as nucleation centers and facilitate CNP formation.
It has been suggested that CNPs have “limited metabolism”20 but we could not detect any metabolic activity in our samples. The presence of acetate has been associated to bacterial growth in the case of NMR spectra obtained from abscesses in vivo29 but in the case of CNPs culture this was discarded due to the lack of 16sRNA in the culture media. Although some authors have reported that CNPs inhibit the PCR reaction,20 it has later been reported that nanobacteria can be easily detected by RT-PCR.30 In our study RT-PCR did not yield any results even when we had evidence of CNP growth.
The hypothesis of the CNP as a living organism is very attractive from a clinical standpoint. According to this hypothesis, antibiotic treatment would have a protective effect on the development of aortic stenosis. All of this is in concordance with prior studies that have led towards the beneficial impact of a combined treatment of EDTA (ethylenediaminetetraacetic acid disodium salt) and tetracyclines in the reduction of calcification at a vascular arterial level.31
Our observations contribute to an alternative interpretation of CNP-induced biomineralization, but do not diminish the importance of further efforts to define the underlying basis of pathological extraskeletal calcification. The concentration of proteins and phosphate in human blood is just below the threshold where hydroxyapatite precipitate starts to form. Factors which facilitate calcium deposition may lead to adverse clinical effects. Therefore, we propose a new theory: protein complexes containing fetuin and albumin normally act as inhibitors of calcification, and may precipitate in the extracellular matrix when this mechanism is overwhelmed. These complexes could then act as nucleation centers for calcium deposition. Recent advances in proteomics and nanotechnology may help in the targeted drug delivery but special attention must be paid in order to avoid the nanostructures being nucleation centers for calcium deposition.32 However, whether fetuin and albumin contribute to the pathogenesis of the disease must be clarified in further studies.
ConclusionsOur results do not support a live origin of CNP. However, due to the ability of CNPs to act as nucleation centers and promote calcification, special attention should be paid to them, particularly in situations when they can be prevented, such as in nanostructure-delivered drugs. Although CNPs are not living entities and cannot be treated as an infection, it cannot be discarded that they are relevant in some diseases in which calcification is a key component.
Conflicts of interestNone declared.
Received 25 November 2011
Accepted 15 March 2012
Corresponding author: Institut de Recerca, Àrea del Cor, Hospital Universitari Vall d’Hebron, Universitat Autònoma de Barcelona, Pg. Vall d’Hebron 119-129, 08035 Barcelona, Spain. ignasi.barba@vhir.org