MicroRNAs (miRNAs) are a class of small noncoding RNA (20-25 nucleotides) involved in gene regulation. In recent years, miRNAs have emerged as a key epigenetic mechanism in the development and physiology of the cardiovascular system. These molecular species regulate basic functions in virtually all cell types, and are therefore directly associated with the pathophysiology of a large number of cardiovascular diseases. Since their relatively recent discovery in extracellular fluids, miRNAs have been studied as potential biomarkers of disease. A wide array of studies have proposed miRNAs as circulating biomarkers of different cardiovascular pathologies (eg, myocardial infarction, coronary heart disease, and heart failure, among others), which may have superior physicochemical and biochemical properties than the conventional protein indicators currently used in clinical practice. In the present review, we provide a brief introduction to the field of miRNAs, paying special attention to their potential clinical application. This includes their possible role as new diagnostic or prognostic biomarkers in cardiovascular disease.
Keywords
Protein-coding nucleotide sequences account for only 1% to 3% of the human genome.1 Originally, the remaining 97% to 99% was considered junk DNA, as it was hypothesized that these sequences did not code biologically relevant information. The number of genes that code for proteins is similar between species with a high and a low degree of biological complexity. Therefore, the degree of biological complexity cannot be explained only by unidirectional gene-to-protein flow2 and may actually be determined by gene expression regulation.3 At present, it is believed that noncoding RNAs (ncRNAs), that is, RNAs not translated into key proteins in the regulation of gene expression, are one of the main sources of this complexity.4 In fact, the percentage of nonprotein-coding genes compared with protein-coding genes is 17-fold higher in humans than the Caenorhabditis elegans5 roundworm.
Noncoding RNAs are classified into 2 subclasses: long ncRNAs (> 200 nucleotides) and short ncRNAs (< 200 nucleotides). MicroRNAs (miRNAs) have a length of 19 to 25 nucleotides and are the smallest RNAs to attract significant attention in recent years. The first miRNA was discovered in 1993 in C. elegans,6,7 but miRNAs were not described in humans until the early 21st century.8 Since then, the field of research has grown exponentially. Like other previously described epigenetic mechanisms, such as DNA methylation or histone modification, miRNAs are important regulators of gene expression that are well-preserved molecules present in all cell types. They are involved in most (if not all) biological processes9 and are a fundamental epigenetic control mechanism at the post-transcriptional level that allows temporal regulation of messenger RNA (mRNA) expression and, therefore, of the rapid response to environmental changes.10 Around 2500 human miRNAs have been identified and catalogued in the miRBase database,11 although recent articles have proposed that the repertoire is actually larger.12
This article provides a brief overview of the biology of intracellular and extracellular miRNAs, as well as a synopsis of the latest information on research related to these molecular species in cardiovascular disease, focusing mainly on their potential as biomarkers for future use in clinical medicine.
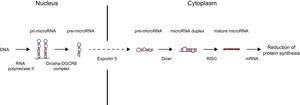
MICRORNA BIOGENESIS AND FUNCTIONDuring their biogenesis, miRNAs follow a multistep process that includes transcription, nuclear maturity, export to cytoplasm, and subsequent processing (Figure).
MicroRNA (miRNA) biogenesis. In the nucleus, a large percentage of miRNA genes are transcribed by RNA polymerase II (a smaller group of these are transcribed by RNA polymerase III). Transcription produces primary miRNA (pri-miRNA) with a length in the hundreds or thousands of nucleotides. In the miRNA canonical biogenesis pathway, the miRNA processing complex (composed of RNase Drosha and DGCR8 cofactor) splices the pri-miRNA, yielding precursor miRNA (pre-miRNA) of 70 to 100 nucleotides in length. This pre-miRNA is exported from the nucleus to the cytoplasm through exportin-5. Once in the cytoplasm, the Dicer enzyme processes and slices pre-miRNA, leading to a small duplex (around 22 nucleotides) containing the mature miRNA and the complementary chain. Each duplex contains 2 mature miRNAs: the guide strand, with a prevalence of 96% to 99%, and the passenger strand. This duplex is associated with a member of the RNA binding protein family (AGO [argonaute]) that assembles the RNA-induced silencing complex (RISC). The guide strand is retained in the complex, whereas the passenger strand is degraded. In some cases, both chains can be functional. Not all known miRNAs have this canonical pathway. In an alternative pathway, miRNAs (known as mirtrons) arise from introns generated during messenger RNA (mRNA) splicing. RISC-miRNA complex binding to mRNA is mediated by Watson-Crick base pairing of the miRNA seed region, a sequence of 6 to 8 nucleotides at the 5’ end of miRNA, with sequences generally complemented in the 3’ untranslated region (UTR) of the target mRNA. Interactions with the coding regions and mRNA 5’ UTR have also been described. Once bound, RISC is able to silence gene expression post-transcriptionally by degradation or inhibition of mRNA translation, leading in both cases to a reduction in the respective target protein levels. The predominant mechanism by which miRNAs reduces protein synthesis is target mRNA deadenylation, a phenomenon that makes mRNA more susceptible to degradation.
In terms of function, miRNAs act at the post-transcriptional level by mRNA degradation or translation inhibition and in both cases, they reduce expression of the gene whose mRNA is targeted. In general, there is only partial complementarity between the nucleotides of the miRNA seed region (a sequence of 6 to 8 nucleotides at the 5’ end) and the target mRNA region. Perfect complementarity between miRNAs and mRNAs is not essential to reduce gene expression and, in fact, rarely occurs in mammals. Both the small size of the seed region and incomplete pairing with the target mRNA allow a single miRNA to suppress the expression of different, often functionally related genes. Moreover, a single mRNA can contain multiple binding sites for various miRNAs and generate a complex network of miRNA-mRNA interactions. It is estimated that miRNAs are able to regulate the expression of approximately 60% of human genes.13 Therefore, miRNAs are a dense network that regulates gene expression with multiple cooperative effects on a large number of genes, making it possible to control various molecular pathways at different levels.2
EXTRACELLULAR MICRORNA: NEW MEDIATORS IN INTERCELLULAR COMMUNICATIONAlthough the first study to report the presence of intact extracellular RNA in the bloodstream was published in 1972,14 the existence of extracellular miRNAs was not proposed until 2007.15 A year later, a key article in the field by Mitchell et al.16 proved the existence of miRNA in blood circulation. Since then, the presence of miRNAs has been described in a large number of body fluids.17
Unlike other types of RNA, such as mRNA, miRNAs are highly stable in the extracellular medium. To protect them from degradation, extracellular miRNAs are packed in different kinds of lipid vesicles,18 such as apoptotic bodies, microparticles, and exosomes, and bound with proteins such as AGO2 (argonaute 2) or NPM1 (nucleophosmin 1)19 or with lipoproteins, including high- and low-density lipoproteins.20 When miRNAs interact with lipid or protein components, they become highly resistant to various harsh conditions, such as degradation due to ribonuclease activity, extreme pH values, high temperatures, repeated freeze-thaw cycles, or prolonged storage.16 In fact, free synthetic miRNAs break down rapidly in blood plasma.16
MicroRNA incorporation in microvesicles occurs selectively.21 Our group has previously shown that the miRNA profile in secreted microvesicles does not necessarily correlate with the intracellular profile in the cell of origin.22 However, there is little information on the cellular machinery involved in miRNA packing inside microvesicles or the protein/lipoprotein-miRNA interaction.20 Likewise, the mechanisms regulating the selection of miRNAs to be exported is still not completely understood. For instance, Villarroya-Beltri et al.23 reported that the cell selects the miRNAs to be exported in exosomes by recognizing specific motifs in the nucleotide sequence. Other authors propose that exosome miRNA content is regulated by overall intracellular miRNA content.24
Extracellular miRNAs act as a genuine intercellular communication system that regulates gene expression and receptor cell phenotype. Like their intracellular forms, extracellular miRNAs participate in both physiological and adaptive responses such as the onset and development of pathologic conditions, including cardiovascular diseases.25,26 Some extracellular miRNAs have already yielded convincing results on their relevance in homeostasis and cardiovascular disease. Zernecke et al.27 proposed that apoptotic bodies generated during atherosclerosis are able to transmit paracrine signals through miRNA by regulating the stability of atheromatous plaque. Hergenreider et al.28 described a mechanism of atheroprotective communication between endothelial cells and vascular smooth muscle cells mediated by extracellular vesicles, miR-143, and miR-145. Shan et al.29 showed that miR-223 is secreted by blood cells and internalized by vascular wall cells and, therefore, plays an important role in the physiology of vascular smooth muscle cells (proliferation, migration, and apoptosis). As a result, they proposed that miR-223 is an endocrine genetic signal between blood cells and vascular cells.
MICRORNA IN CARDIOVASCULAR PHYSIOLOGY AND PATHOPHYSIOLOGYAbnormal miRNA expression levels directly affect expression of the respective target mRNAs and, therefore, miRNAs are potentially causative elements of disease. The first report described the role of miRNAs in the development of chronic leukemia,30 but numerous subsequent studies have correlated changes to miRNA expression profiles with the onset and progression of various illnesses, including cancer, metabolic diseases, and neurological disorders.31 The relevance of miRNA gene expression regulation in cardiovascular development and homeostasis as well as the implications of miRNAs for cardiovascular disease are well established.32 Excellent reviews have been published on the role of miRNAs in some aspects related to functionality and various cardiovascular conditions.33–35
Abnormal expression profiles in the different tissues that comprise the cardiovascular system are associated with disorders such as heart failure, atherosclerosis, and cardiomyopathies of different etiology.36 miRNAs are expressed as cardiomyocytes, fibroblasts, endothelial cells, and vascular smooth muscle cells and control virtually all aspects of cardiovascular biology, including cardiac remodeling and fibrosis, apoptosis, inflammation, proliferation, angiogenesis, and metabolism. In this regard, various studies have shown that miRNA-specific overexpression and repression in in vitro and in vivo models including miR-1, miR-126, miR-133a, or miR-208a, among others, are key mechanisms in cardiovascular disease.36,37 A good example of the role of miRNAs in cardiovascular development and pathophysiology has been obtained with the conditional knockout mouse model for Dicer, a cardiac enzyme involved in miRNA biogenesis. This mouse model includes dilated cardiomyopathy and heart failure,38 as well as congenital cardiovascular defects.39
CIRCULATING MICRORNA AS BIOMARKERS OF CARDIOVASCULAR DISEASEThe identification of participants in subclinical stages of disease or at high cardiovascular risk has crucial implications for patient health, as well as for efficient management of health system resources, particularly in the current context of the high prevalences of mortality and morbidity associated with cardiovascular disease. Therefore, it is important to discover new biomarkers and to improve diagnostic tools, particularly for risk prediction algorithms.40,41
Circulating miRNAs have adequate biological and physicochemical properties to be useful biomarkers in clinical practice3: a) they can be obtained by minimally invasive techniques in the types of samples currently used in clinical laboratories; b) they can arise from necrotic cells or be actively secreted from live cells; c) circulating miRNA profiles may show high specificity according to tissue and disease; d) circulating miRNA profiles are affected in situations of cellular stress and in pathophysiologic conditions; e) miRNAs are highly stable and have a long half-life in the sample; f) like nucleic acids, miRNAs have advantages over biomarkers presently used in clinical practice: peptide-based biomarkers may have different variants of the same molecule and be subject to post-translational changes that hinder their detection. Due to their small size and chemical composition, miRNAs are less complex molecules than most biological molecules in plasma, simplifying assays; g) miRNAs can be quantified efficiently, profitably, and relatively rapidly in current clinical laboratories with high sensitivity and specificity using real-time polymerase chain reaction, and h) global profiles can be obtained in a single experiment by real-time polymerase chain reaction or readily accessible techniques such as sequencing or microarrays.
Because of these characteristics, several authors have proposed that miRNA assays be used clinically in the short or medium term.42 An miRNA-based blood test could be a potent noninvasive tool that would provide clinicians with valuable information for adequate patient management.
The role of miRNAs as biomarkers in some of the clinical entities that fall under the term cardiovascular disease will be addressed below, although covering all conditions and all miRNAs proposed as biomarkers is beyond the scope of this article.
Acute Coronary SyndromeOne of the main fields explored when investigating miRNAs as cardiovascular disease biomarkers is their diagnostic potential in acute coronary syndrome (Table).
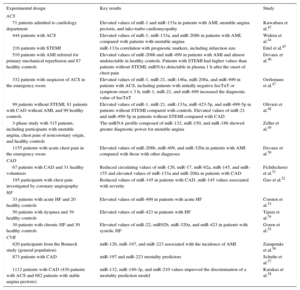
Selection of Studies on Circulating MicroRNA and Cardiovascular Disease
| Experimental design | Key results | Study |
|---|---|---|
| ACS | ||
| 71 patients admitted to cardiology department | Elevated values of miR-1 and miR-133a in patients with AMI, unstable angina pectoris, and tako-tsubo cardiomyopathy | Kuwabara et al.43 |
| 444 patients with ACS | Elevated values of miR-1, miR-133a, and miR-208b in patients with AMI, compared with patients with unstable angina | Widera et al.44 |
| 216 patients with STEMI | miR-133a correlation with prognostic markers, including infarction size | Eitel et al.45 |
| 510 patients with AMI referred for primary mechanical reperfusion and 87 healthy controls | Elevated values of miR-208b and miR-499 in patients with AMI and almost undetectable in healthy controls. Patients with STEMI had higher values than patients without STEMI; miRNAs detectable in plasma 1 h after the onset of chest pain | Devaux et al.46 |
| 332 patients with suspicion of ACS in the emergency room | Elevated values of miR-1, miR-21, miR-146a, miR-208a, and miR-499 in patients with ACS, including patients with initially negative hscTnT or symptom onset < 3 h; miR-1, miR-21, and miR-499 increased the diagnostic value of hscTnT | Oerlemans et al.47 |
| 99 patients without STEMI, 81 patients with CAD without AMI, and 99 healthy controls | Elevated values of miR-1, miR-21, miR-133a, miR-423-5p, and miR-499-5p in patients without STEMI compared with controls. Elevated values of miR-21 and miR-499-5p in patients without STEMI compared with CAD | Olivieri et al.48 |
| 3-phase study with 315 patients, including participants with unstable angina, chest pain of noncoronary origin, and healthy controls | The miRNA profile composed of miR-132, miR-150, and miR-186 showed greater diagnostic power for unstable angina | Zeller et al.49 |
| 1155 patients with acute chest pain in the emergency room | Elevated values of miR-208b, miR-499, and miR-320a in patients with AMI compared with those with other diagnoses | Devaux et al.50 |
| CAD | ||
| 67 patients with CAD and 31 healthy volunteers | Reduced circulating values of miR-126, miR-17, miR-92a, miR-145, and miR-155 and elevated values of miR-133a and miR-208a in patients with CAD | Fichtlscherer et al.51 |
| 195 participants with chest pain investigated by coronary angiography | Reduced values of miR-145 in patients with CAD. miR-145 values associated with severity | Gao et al.52 |
| HF | ||
| 33 patients with acute HF and 20 healthy controls | Elevated values of miR-499 in patients with acute HF | Corsten et al.53 |
| 50 patients with dyspnea and 39 healthy controls | Elevated values of miR-423 in patients with HF | Tijsen et al.54 |
| 30 patients with chronic HF and 30 healthy controls | Elevated values of miR-22, miR92b, miR-320a, and miR-423 in patients with systolic HF | Goren et al.55 |
| CVR | ||
| 820 participants from the Bruneck study (general population) | miR-126, miR-197, and miR-223 associated with the incidence of AMI | Zampetaki et al.56 |
| 873 patients with CAD | miR-197 and miR-223 mortality predictors | Schulte et al.57 |
| 1112 patients with CAD (430 patients with ACS and 682 patients with stable angina pectoris) | miR-132, miR-140-3p, and miR-210 values improved the discrimination of a mortality prediction model | Karakas et al.58 |
ACS, acute coronary syndrome; AMI, acute myocardial infarction; CAD, coronary artery disease; CVR, cardiovascular risk; HF, heart failure; hscTnT, highly-sensitive cardiac troponin T; miRNA, microRNA; STEMI, ST-segment elevation myocardial infarction.
The vast majority of studies focus on miRNA-rich plasma levels in myocardial tissue (miR-1, miR-133a/b, miR-208a, and miR-499). Skeletal muscle also have high levels of miRNAs, known as myomiRs. The plasma concentration of these miRNAs rises shortly after myocardial necrosis, with similar kinetics to that observed with conventional cardiac indicators, such as highly-sensitive cardiac troponin T.43,45,46 miRNA values are increased in the transcoronary gradients of patients with acute coronary syndrome, suggesting that miRNAs are released from necrotic cardiomyocytes.59 In fact, a meta-analysis by Cheng et al.60 identified miR-133a and miR-499 as potential clinical biomarkers of myocardial infarction. Other miRNAs (miR-21, miR-92, miR-126, and miR-132) have been proposed as indicators with high sensitivity and specificity not for only myocardial infarction, but also for unstable angina.49,50,61,62 Circulating miRNAs could provide added diagnostic value to cardiac troponins.47 In particular, the potential of circulating miRNAs as biomarkers has been demonstrated in studies that analyze their capacity to improve diagnosis in patients with chest pain.47 For instance, Zeller et al.49 proposed that the combination of miR-132, miR-150, and miR-186 has superior diagnostic value for unstable angina than classic combined indicators, such as cardiac troponin I, B-type natriuretic peptide, and C-reactive protein.
The capacity of circulating miRNAs to provide information in the diagnosis of unstable and stable angina,63 myocardial infarction and unstable angina,44 and non–ST-segment elevation acute myocardial infarction and acute heart failure48 has also been reported in several studies.
Coronary DiseasePlasma proteins such as fibrinogen or C-reactive protein have been proposed as biomarkers for coronary disease.64 However, the diagnostic value of these indicators is limited because they are modulated by factors not necessarily related to cardiovascular disease.64 Currently available imaging techniques do not allow early screening for the disease. Consequently, it is essential to develop sufficiently robust new biomarkers to identify and characterize atherosclerotic lesions, particularly those that are unstable.65
Various studies have analyzed the association of circulating miRNAs with coronary disease (Table). In a pioneer study, Fichtlscherer et al.51 proposed circulating values of miR-126, miR-17, miR-92a, miR-133a, miR-145, miR-155, and miR-208a as indicators of the presence of coronary disease. Some of these miRNAs have been subsequently validated in independent studies.66 The circulating miRNA profile appears to be associated not only with the presence of coronary lesion, but also with the severity of the illness.52,66,67 miRNAs have also been proposed as potential biomarkers for vulnerable plaques.62 Many of these results were obtained in case-control studies with small populations, however.
Heart FailureVarious studies have shown a specific circulating miRNA profile in patients with heart failure (Table). Marfella et al.68 evaluated a panel of 84 miRNAs previously associated with structural abnormalities of the heart, finding a circulating profile of 24 miRNAs characteristic of patients with heart failure. Apart from some exceptions,69 several studies mention miR-423 as a potential biomarker associated with heart failure. In one of the earliest studies in this field, Tijsen et al.54 identified 6 miRNAs that are elevated in patients with heart failure, among which miR-423 showed a strong association with a clinical diagnosis of the disease. Later studies have validated these results.55 Various studies have suggested that miRNAs may be novel biomarkers of heart failure with normal ejection fraction,70 diastolic dysfunction,71 and acute heart failure.53
Cardiovascular RiskCardiovascular risk is currently evaluated exclusively on the basis of established classic risk factors, such as hypertension, dyslipidemia, diabetes, and smoking. Unfortunately, these traditional risk factors do not fully explain the risk of a cardiovascular event. Many of the events occur in patients at low-to-intermediate risk with only a few of the classic cardiovascular risk factors.65 In contrast, many individuals classified as high risk according to these factors do not experience any cardiovascular event; not even in the long-term.72 Consequently, there is clear clinical interest in the development of novel noninvasive biomarkers that are readily accessible and significantly improve the predictive capacity of the algorithms developed to date, beyond the traditional risk factors.3
Various studies have analyzed the capacity of circulating miRNAs to predict future cardiovascular events in both the general population and in patients with known coronary disease. Zampetaki et al.56 first demonstrated the potential of miRNAs to predict adverse events. These authors followed a cohort of 820 individuals for 10 years, finding a significant association between 3 miRNAs (miR-126, miR-197, and miR-223) and the risk of myocardial infarction, even after adjusting for possible confounding factors. The prognostic potential of miR-197 and miR-223 as predictors of cardiovascular mortality was later corroborated in a cohort of 873 patients with coronary disease.57 Additionally, miR-132, miR-140, and miR-210 were also associated with cardiovascular death in a similar cohort of 1112 patients.58 Bye et al.73 recently proposed that a panel of 5 miRNAs (miR-106a-5p, miR-424-5p, let-7g-5p, miR-144-3p, and miR-660-5p) is able to improve myocardial infarction risk prediction in healthy individuals. These results show the promising applicability of miRNAs as biomarkers in primary and secondary prevention of cardiovascular disease.
Other Cardiovascular DiseasesCirculating miRNAs have been addressed by only a few of the studies investigating them as biomarkers of other cardiovascular diseases, beyond those mentioned. In this regard, only a few investigations have analyzed the association between circulating miRNAs and stroke, despite its high incidence. An analysis of miRNA expression in the peripheral blood of young patients with ischemic stroke revealed that miRNA peripheral blood profiles may be potential biomarkers in the diagnosis of cerebral ischemic stroke.74 The blood miRNA profile can also discriminate between hemorrhagic and ischemic stroke.75 Other authors have shown that serum miRNAs are clinically useful indicators in abdominal aortic aneurysm.76 The potential role of miRNAs as biomarkers of peripheral atherosclerosis has also been evaluated. Stather et al.77 identified a specific peripheral arterial disease profile that showed good diagnostic potential and also provided information on possible molecular pathways involved in the pathogenesis of the disease. Recently, a detailed review looked at the possible role of circulating miRNAs as biomarkers for different cardiovascular disorders, including atrial fibrillation, myocarditis, pulmonary embolism, tako-tsubo cardiomyopathy, diabetes-related vascular complications, and vascular dementia.78,79
LIMITATIONS IN THE CLINICAL APPLICATION OF CIRCULATING MICRORNA AS BIOMARKERSDespite the promising results of circulating miRNAs as novel cardiovascular disease biomarkers, miRNAs have still not been incorporated in clinical practice, mainly due to the lack of large cohort studies and the technical limitations of their application.9,37 Existing studies also show considerable heterogeneity, including differences related to the miRNAs and the diseases studied, as well as the experimental designs, sample collection and processing, and methodologies used to assay miRNAs.80 For instance, a large number of published studies have not compared the diagnostic or prognostic potential of circulating miRNAs with previously established clinical indicators currently used in real-life clinical practice, even though this would be a key step in the development of new biomarkers. Furthermore, selection of controls has been inadequate, particularly with regard to confounding factors such as age, sex, medication, and comorbidities.81 Another limitation is the lack of specificity of the miRNAs proposed as biomarkers. For example, miR-126 has been strongly associated with coronary disease.51 However, this same miRNA is also closely linked to heart failure82 and diabetes,83 raising questions about its clinical usefulness.
The implementation of miRNA-based diagnoses in routine clinical practice has also been hampered by technical difficulties. Standard methods have not yet been established to isolate and quantify circulating miRNAs, hampering comparison of studies undertaken by different laboratories. Likewise, current miRNA screening techniques do not allow rapid diagnosis, despite this often being necessary in patients with acute myocardial infarction, for instance.
OTHER EPIGENETIC CIRCULATING BIOMARKERS OF CARDIOVASCULAR DISEASEApproaches based on ncRNAs provide new opportunities to develop novel biomarkers. In recent years, in addition to miRNAs, a number of studies propose using circulating long ncRNAs (> 200 nucleotides) as sensitive and specific biomarkers not only of cardiovascular disease,84,85 but also of therapeutic response.86 The clinical application of these species is more advanced in the field of oncology, where tests have been developed to screen urine for the long ncRNA known as PCA3, a biomarker highly specific for prostate cancer.87
Other epigenetic mechanisms, such as DNA methylation or histone modification, could be a very useful source of circulating biomarkers. However, very few studies have investigated the role of these epigenetic mechanisms as useful indicators in clinical practice. A review has recently been published of the association between DNA methylation and the presence of cardiovascular disease.88 In fact, hypomethylation of the LINE-1 sequence in peripheral blood cells or visceral adipose tissue is associated with diabetes, obesity, lower high-density lipoprotein cholesterol, and elevated values of total cholesterol and inflammation, even independently of other risk factors.89,90
FUTURE PROSPECTSThe first extracellular miRNAs were described only a decade ago. Although relatively little time has elapsed since then, we have witnessed considerable advances in their possible clinical application as biomarkers. The scientific community has great expectations for the clinical use of circulating miRNAs as diagnostic and prognostic tools of cardiovascular disease. The fact that they can be readily detected by the methodology currently used in clinical laboratories and are associated with cardiovascular conditions (in some cases, superior to that observed with conventional indicators) suggests their potential value as biomarkers. However, several aspects should be considered before miRNAs are established as new clinical tools. In coming years, a key step will be replication of these results in independent multicenter studies with large populations to elucidate the real-life clinical applicability of circulating miRNAs.
FUNDINGOur study received aid from the Carlos III Health Institute, which is cofunded with financial assistance from the ERDF (European Regional Development Fund) (V. Llorente-Cortés: FIS PI14/01729), the Carlos III Health Institute (Cardiovascular Biomedical Research Network Center: CB16/11/00403), the TV3 Marathon Foundation (V. Llorente-Cortes: 201521-10) and the Carlos III Health Institute, Ministry of Economy and Competitiveness (E. Iglesias-Gutiérrez: DEP2012-39262). D. de Gonzalo-Calvo has a Sara Borrell contract (CD14/00109) with the Carlos III Health Institute. D. de Gonzalo-Calvo and V. Llorente-Cortés are members of the European network Cardiolinc.
CONFLICTS OF INTERESTNone declared.

![MicroRNA (miRNA) biogenesis. In the nucleus, a large percentage of miRNA genes are transcribed by RNA polymerase II (a smaller group of these are transcribed by RNA polymerase III). Transcription produces primary miRNA (pri-miRNA) with a length in the hundreds or thousands of nucleotides. In the miRNA canonical biogenesis pathway, the miRNA processing complex (composed of RNase Drosha and DGCR8 cofactor) splices the pri-miRNA, yielding precursor miRNA (pre-miRNA) of 70 to 100 nucleotides in length. This pre-miRNA is exported from the nucleus to the cytoplasm through exportin-5. Once in the cytoplasm, the Dicer enzyme processes and slices pre-miRNA, leading to a small duplex (around 22 nucleotides) containing the mature miRNA and the complementary chain. Each duplex contains 2 mature miRNAs: the guide strand, with a prevalence of 96% to 99%, and the passenger strand. This duplex is associated with a member of the RNA binding protein family (AGO [argonaute]) that assembles the RNA-induced silencing complex (RISC). The guide strand is retained in the complex, whereas the passenger strand is degraded. In some cases, both chains can be functional. Not all known miRNAs have this canonical pathway. In an alternative pathway, miRNAs (known as mirtrons) arise from introns generated during messenger RNA (mRNA) splicing. RISC-miRNA complex binding to mRNA is mediated by Watson-Crick base pairing of the miRNA seed region, a sequence of 6 to 8 nucleotides at the 5’ end of miRNA, with sequences generally complemented in the 3’ untranslated region (UTR) of the target mRNA. Interactions with the coding regions and mRNA 5’ UTR have also been described. Once bound, RISC is able to silence gene expression post-transcriptionally by degradation or inhibition of mRNA translation, leading in both cases to a reduction in the respective target protein levels. The predominant mechanism by which miRNAs reduces protein synthesis is target mRNA deadenylation, a phenomenon that makes mRNA more susceptible to degradation. MicroRNA (miRNA) biogenesis. In the nucleus, a large percentage of miRNA genes are transcribed by RNA polymerase II (a smaller group of these are transcribed by RNA polymerase III). Transcription produces primary miRNA (pri-miRNA) with a length in the hundreds or thousands of nucleotides. In the miRNA canonical biogenesis pathway, the miRNA processing complex (composed of RNase Drosha and DGCR8 cofactor) splices the pri-miRNA, yielding precursor miRNA (pre-miRNA) of 70 to 100 nucleotides in length. This pre-miRNA is exported from the nucleus to the cytoplasm through exportin-5. Once in the cytoplasm, the Dicer enzyme processes and slices pre-miRNA, leading to a small duplex (around 22 nucleotides) containing the mature miRNA and the complementary chain. Each duplex contains 2 mature miRNAs: the guide strand, with a prevalence of 96% to 99%, and the passenger strand. This duplex is associated with a member of the RNA binding protein family (AGO [argonaute]) that assembles the RNA-induced silencing complex (RISC). The guide strand is retained in the complex, whereas the passenger strand is degraded. In some cases, both chains can be functional. Not all known miRNAs have this canonical pathway. In an alternative pathway, miRNAs (known as mirtrons) arise from introns generated during messenger RNA (mRNA) splicing. RISC-miRNA complex binding to mRNA is mediated by Watson-Crick base pairing of the miRNA seed region, a sequence of 6 to 8 nucleotides at the 5’ end of miRNA, with sequences generally complemented in the 3’ untranslated region (UTR) of the target mRNA. Interactions with the coding regions and mRNA 5’ UTR have also been described. Once bound, RISC is able to silence gene expression post-transcriptionally by degradation or inhibition of mRNA translation, leading in both cases to a reduction in the respective target protein levels. The predominant mechanism by which miRNAs reduces protein synthesis is target mRNA deadenylation, a phenomenon that makes mRNA more susceptible to degradation.](https://static.elsevier.es/multimedia/18855857/0000007000000009/v1_201708280843/S1885585717302815/v1_201708280843/en/main.assets/thumbnail/gr1.jpeg?xkr=eyJpdiI6IldBdEYvVHJ5cHhjREJJSDBXbDlML1E9PSIsInZhbHVlIjoicnBkQVNneVJpcHNhUDlPaGhkYTJHWFVIMkx0SkFXSnQyVmhUSVhySXc3ND0iLCJtYWMiOiJiNmE5ZGRhMzI5NGJjYzg1NDQ3YWY2NWIxNjhiMTAyZGJhNDcyMjgyNzYzN2ZjMjkxY2UwNGVmZmQ0ZWVkMmNkIiwidGFnIjoiIn0=)