The observation that “one size does not fit all” for the prevention and treatment of cardiovascular disease, among other diseases, has driven the concept of precision medicine. The goal of precision medicine is to provide the best-targeted interventions tailored to an individual's genome. The human genome is composed of billions of sequence arrangements containing a code that controls how genes are expressed. This code depends on other nonstatic regulators that surround the DNA and constitute the epigenome. Moreover, environmental factors also play an important role in this complex regulation. This review provides a general perspective on the basic concepts of molecular biology related to genetics and epigenetics and a glossary of key terms. Several examples are given of polymorphisms and genetic risk scores related to cardiovascular risk. Likewise, an overview is presented of the main epigenetic regulators, including DNA methylation, methylcytosine-phosphate-guanine-binding proteins, histone modifications, other histone regulations, micro-RNA effects, and additional emerging regulators. One of the greatest challenges is to understand how environmental factors (diet, physical activity, smoking, etc.) could alter the epigenome, resulting in healthy or unhealthy cardiovascular phenotypes. We discuss some gene-environment interactions and provide a methodological overview.
Keywords
Technological advances made during the Human Genome Project and after its completion have greatly reduced costs and increased the immediacy of access to genetic test results.1 Genetic markers can now be determined as part of routine investigation of large patient cohorts2,3 and clinical trials.4,5 The fruits of decades of research into genetic markers have deepened our understanding of the molecular bases of the most common diseases, including cardiovascular diseases and their intermediate phenotypes.6,7 However, the huge amount of information on new genes linked to the various diseases and their possible environmental modulation remains in the theoretical realm and subsequent diagnostic, preventive, and therapeutic validation is required before these markers can be translated to clinical practice and applied to the general population. To accelerate this process, an initiative called precision medicine was launched in 2015 in the United States.8 As detailed by Collins and Varmus in their 2015 publication on this approach,8 the concept of precision medicine—defined as “the implementation of prevention and treatment strategies that take individual variability into account to optimize results”—is not new and has been used in one form or another for many years, such as when patients are blood typed to guide blood transfusions. However, the novelty lies in the huge number of new genetic markers that can help to improve our understanding of patients’ genetic risk and predict interindividual treatment responses. In addition to genetic markers based on DNA sequences, numerous other -omic biomarkers are available (eg, epigenomic, transcriptomic, metabolomic, proteomic). In conjunction with bioinformatics and new computational tools for the handling and integration of these data, these biomarkers can provide highly valuable information that markedly improves disease prevention and treatment. Although initial efforts in precision medicine have largely focused on cancer,8 advances are also being made in the field of cardiovascular disease that will allow goals to be reached in this new era of medicine.9
For precision medicine to move from dream to reality,10 sustained research effort is needed to incorporate information gleaned on -omic markers into clinical trials and other epidemiological studies and generate results with a high level of scientific evidence to guide decisions in the new era.8 This step requires medical and other biomedical scientific professionals to acquire a solid knowledge base in -omics in order to better critically interpret and confront the new challenges. Although the central dogma of biology, proposed by Francis Crick in 1958, proposed a unidirectional flow of the transmission and expression of genetic information, namely, transcription of DNA into messenger RNA and translation of messenger RNA into protein, the factor that finally performs the cellular action,11 it is now known that this pathway is not universal and that powerful regulatory elements can allow the same DNA to give rise to 2 or 3 distinct proteins. Greater understanding is thus required of not only the genetic elements, but also the epigenetic ones contributing to this regulation. Accordingly, the aim of this work is to provide a succinct and up-to-date overview of the basic molecular biology concepts related to genetics and epigenetics. Our approach involves various examples of the studies performed, focusing on cardiovascular diseases, and culminating in a methodological reflection on what are known as gene-environment interactions. Other medical journals such as the New England Journal of Medicine12,13 and Journal of the American Medical Association,14–16 as well as more specialized publications,17–20 have also published reviews of the basic concepts in genetics and epigenetics and should be consulted to complement this work.
THE HUMAN GENOMEThe human genome contains about 6 billion DNA base pairs (adenine, thymine, guanine, and cytosine) and is organized into 23 pairs of chromosomes. There are an estimated 20 000 to 25 000 genes in the human genome, much fewer than originally predicted,12 partly because the same gene can give rise to various proteins,13 as described below. Because each DNA base pair is about 0.34nm in length, each diploid cell would contain about 2 meters of DNA if it were fully uncoiled. In total, the human body would contain 100 billion meters of DNA.21 Histones are in charge of compacting the DNA so that it can fit within the microscopic cell nucleus. Histones are a family of small positively charged proteins named H1, H2A, H2B, H3, and H4.22 Because DNA is negatively charged, due to the phosphate groups in the phosphate-sugar backbone, histones strongly bind to DNA. The basic structural and functional unit of chromatin is the nucleosome, which contains 8 histone proteins and about 146 DNA base pairs. Nucleosomes in turn form part of another structure called the chromatosome, with each chromatosome packaging an average 100 million base pairs. Thus, each chromosome is a long chain of nucleosomes.21
Chromatin is in turn classified into euchromatin and heterochromatin. These 2 forms show differences in staining, structure, and function but euchromatin is basically a more relaxed and transcriptionally active structure that encompasses most genes, whereas heterochromatin is denser and contains more repeat sequences, such as those found in telomeres. The latter are highly repetitive regions located at the end of chromosomes. The most repeated sequence in human telomeres is 5’-TTAGGG-3’, with sometimes even more than 2000 repeats. There is a complex of 6 proteins associated with telomeres called shelterin or telomere protection complex, composed of TRF1 and TRF2, that in turn interacts with RAP1, TPP1, POT1, and TIN2 to associate with telomeric DNA.22 The enzyme in charge of telomere extension is called telomerase. This reverse transcriptase maintains telomere length; using RNA as a template and in conjunction with specific accessory proteins, it repeatedly adds the sequence d(TTAGGG) at the extreme 3’ end of telomeric DNA.22 Various studies have associated telomere length with different diseases.23 It is generally believed that shorter telomeres are linked to greater aging and higher cardiovascular risk.24
Human Genome Sequence VariationsGenes constitute the transcriptionally active part of chromosomes. The basic structure of a gene is divided into introns and exons.19 Introns are noncoding sequences, whereas exons are coding. There are also noncoding regions at the start and end of each gene called the 5’-untranslated region (UTR) and 3’-UTR. The initial region contains the gene promoter and the end region is highly involved in regulation by microRNA. The DNA sequences can be subject to variations of different types (eg, insertions, deletions, repeat expansions).14,19 The best known are single nucleotide changes, more commonly known by the abbreviation SNP (single nucleotide polymorphism).14 These SNPs can be found in all gene regions. Polymorphisms found in introns were once thought to have no effect on the amino acid sequence of the resulting protein and were said to be nonfunctional. However, the significance of polymorphisms in noncoding regions is now better understood: such variations can actually affect gene functionality without the involvement of an amino acid change, with or without the binding of other regulators, as discussed below. Polymorphisms found in exons do not always cause amino acid changes because, even if there is a base change, the corresponding triplet (group of 3 nucleotides determining an amino acid) that is generated can still code for the same amino acid due to the “degenerate” nature of the genetic code. Accordingly, if the combinations of the 4 elements (4 DNA bases) and 3 bases per codon are counted, they would code for 64 distinct amino acids. However, there are only 20 different amino acids because, as observed by Wittmann in 1962, an amino acid can be encoded by more than 1 codon.
In quantitative terms, 2 unrelated people are estimated to share more than 99% of DNA sequences but, taking into account the billions of base pairs forming the genome, the DNA sequences of 2 unrelated people can vary by over 20 million bases.12,13 Some of these base changes can increase the risk of disease, as happens in the types of disorder known as monogenic diseases. These conditions show high penetrance, with a single base change in a specific DNA region sufficient to give rise to a well-characterized disease. Various types of phenotypes have been linked to cardiovascular diseases with a monogenic origin, such as monogenic familial hypercholesterolemia caused by one or more mutations in the low-density lipoprotein receptor gene.25 A monogenic component is also the main cause of certain heart diseases, some of which are congenital.26 Despite the high penetrance of monogenic diseases, their frequency in the population is very low. Typically, the main cardiovascular diseases and their intermediate phenotypes respond to a polygenic inheritance pattern involving various types of polymorphisms, with each having a small effect that slightly increases the risk. A glossary of key terms related to genetics and epigenetics is presented in the Table. Understanding of the significance of these terms is required to better comprehend the concepts discussed in this article.
Glossary of Terms in Genetics and Epigenetics
| Term | Description |
|---|---|
| Allele | Each one of the versions of a polymorphism or gene. An individual typically inherits 2 alleles for each polymorphism, 1 from each parent. Individuals with 2 equal alleles are homozygous for that polymorphism. They are heterozygous if the alleles are different. |
| Autosome | One of the nonsex chromosomes |
| Chromatin | Material formed by nucleic acids and proteins seen in the cell nucleus during interphase |
| Chromosome | Organized package of DNA found in the cell nucleus. Each organism has a different number of chromosomes. Humans have 23 pairs of chromosomes |
| Codon | A sequence of 3 DNA or RNA bases that specifies a single amino acid in the translation process |
| DNA | The chemical name of the molecule containing the genetic instructions. It consists of 2 strands that are wound around each other to form a double helix. Bound to each sugar is 1 of 4 bases: adenine (A), cytosine (C), guanine (G), and thymine (T). The 2 strands are bound to each other via well-defined links between specific bases |
| Epigenetic | Mechanism of the regulation (transcription and translation) of gene expression that does not rely on changes in DNA bases but that operates at a higher level |
| EWA | Epigenome-wide association. It refers to studies analyzing the methylation of the whole epigenome |
| Exon | Coding region of a gene |
| Gene | Unit of inheritance that occupies a specific position in the genome (locus) and has a particular structure |
| Genome | Basic set of chromosomes containing all genetic information of an individual |
| Genotype | Sum of an individual's alleles for a specific position |
| GRS | Genetic risk score. This score can be weighted or unweighted depending on whether the effect size of each polymorphism is evaluated or only the number of risk alleles |
| GWAS | Genome-wide association study |
| Histone | Small basic pH proteins, rich in lysine and arginine, that are bound to DNA at the chromatin |
| Locus | Place occupied by a gene in the genome |
| Methylation | Addition of methyl groups (-CH3) to DNA in the form of methylated bases |
| MicroRNA | Tiny fragment of noncoding RNA with an important regulatory function |
| Mutation | Any base change introduced in the DNA sequence. Sometimes specifically used to indicate that the allelic frequency of a genetic variation is very low (< 1%) |
| Nucleotide | Molecule formed by a nitrogenous base, a pentose sugar, and a phosphoric acid group. Basic unit of nucleic acid |
| Phenotype | Characteristic or observable trait of individuals, resulting from an interaction between their genotype and the environment in which it is expressed. Final cardiovascular disease phenotypes (eg, infarction, stroke) are distinguished from intermediate phenotypes (eg, hypertension, dyslipidemias) |
| RNA | Molecule formed by a polyribonucleotide of varying lengths that contains uracil instead of thymine. There are 3 types: messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA) |
| Telomere | Extreme end of chromosomes. In humans, the DNA of telomeres is composed of tandem repeats of the sequence TTAGGG |
In recent years, spectacular advances have been made in the technology used to determine DNA variations. Highly accurate, fast, and inexpensive data can now be obtained on the presence of specific genetic variants in an individual's genome. The economic cost and time required depend on the number of genetic variations to be analyzed. The first step in the genome analysis process involves DNA extraction. To analyze genomic DNA, any biological sample containing nucleated cells can be used. Peripheral blood leukocytes are the standard specimen. The DNA extracted from this source is of a sufficient concentration and quality for most subsequent genetic analyses. As a noninvasive alternative for biological sample collection, saliva can be used, but the concentration and quality of the DNA extracted from this specimen might be insufficient for massive genetic analyses, which is why researchers must weigh up the pros and cons of each option.
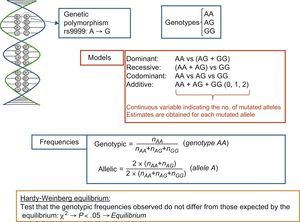
If the goal is simply to analyze a single polymorphism or a small number in candidate genes, the SNP determination technique used is typically based on fluorescent probes; this technique replaces an earlier one involving restriction enzymes and agarose gels, known as restriction fragment length polymorphism analysis. For polymorphism nomenclature, a system has been adopted to unequivocally identify each genetic variant. In this system, polymorphisms are assigned a number with the prefix “rs” (“reference SNP”).14 This is an opportune moment to clarify the conceptual difference between mutations and polymorphisms. The difference is academic because the terms are frequently used synonymously. Both refer to DNA base changes, and the difference lies in their frequency. The term mutation refers to uncommon variants (with an allelic frequency < 1%), whereas polymorphism refers to variations with allelic frequencies > 1%. An example of a DNA variant consisting of an A>G change (rs9999) is shown in Figure 1. This change causes 2 alleles (allele A and allele G). These alleles give rise to 3 corresponding genotypes and distinct inheritance models (dominant, recessive, codominant, and additive) can be used when studying their association with disease phenotypes. The figure also shows how to calculate allelic and genotype frequencies and illustrates the Hardy-Weinberg equilibrium test.
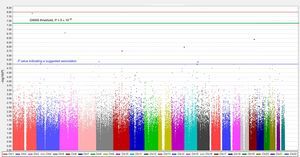
To study a greater number of polymorphisms, distinct types of chips are used allowing higher-density genotyping. These chips include those termed “whole-genome” chips, thus named because they include polymorphisms distributed throughout all chromosomes. The first whole-genome genotyping chips contained 10 000 SNPs and were also called 10K chips (with K indicating thousands). Chip density has since increased. Thus, for example, the first genome-wide association studies (GWASs) performed in the Framingham study, published in 2007, used 100K genotyping chips.27 The density of these chips has subsequently increased even further, and current chips have genotyping densities of more than 1 million SNPs per individual. Statistical analyses of the association between whole-genome genotyping and the cardiovascular phenotype of interest28 frequently use graphs called Manhattan plots, due to their similarity to the skyscrapers of Manhattan Island in New York. Figure 2 shows a Manhattan plot corresponding to a dense GWAS using a 1000K array and an intermediate cardiovascular phenotype. The vertical axis is the–log10 value of the P for association between each SNP and the phenotype of interest, whereas the horizontal axis is the position occupied by each SNP in the chromosome. Each point on the graph is an SNP, so that only those SNPs with very low P values can be seen; the other SNPs are visible as a denser mass in the lower part of the graph. The higher the position of the SNP, the stronger its association with the cardiovascular phenotype (ie, with a lower P value for association). For an association to be considered statistically significant, a nominal P value less than .05 is not used. Instead, this figure is corrected by the number of comparisons made to minimize the occurrence of false positives. The commonly accepted threshold value for a statistically significant association in GWASs is P < 5 × 10–8; this P value is equivalent to a –log value of 7.25. Some authors also use P < 10–8, which is equivalent to a –log value of 8. When an exploratory GWAS is performed, whose results are to be confirmed in another population, the P values determined are used as suggestive association results for subsequent confirmation. This value is P < 10–5. Hundreds of GWASs have been performed with intermediate and final cardiovascular disease phenotypes and the main genes and their corresponding SNPs have been identified for each disease. Given that a detailed list of these polymorphisms is beyond the scope of this review, we recommend that the original publications be consulted or the relevant synthetic reviews.6,28
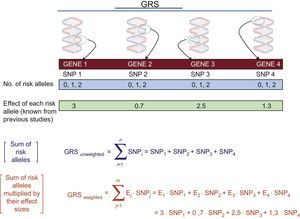
The GWASs enable us to individually determine the main SNPs associated with the phenotype of interest. To determine their joint contribution, genetic risk scores (GRSs) are used. Figure 3 shows the calculation of the 2 main types of GRSs: a) unweighted and b) weighted. Numerous studies have analyzed and quantified the influence of various GRSs associated with distinct cardiovascular phenotypes.29 Greater detail on the calculation of GRSs and their advantages and disadvantages can be found in other reviews.30
At the same time, there has also been a considerable leap forward in direct sequencing technology, now known as next-generation sequencing (NGS). This approach, instead of being based on techniques applied in the Human Genome Project (the traditional Sanger capillary electrophoresis sequencing method, considered the first generation), NGS techniques obtain more data at lower cost. The NGS has 3 major improvements31: a) it does not require a bacterial cloning step and sequencing libraries are prepared in a cell-free system; b) millions of sequencing reactions are processed in parallel and at the same time; c) base detection is performed cyclically and in parallel. All of these factors greatly improve reliability and reduce cost. Various projects are underway that aim to sequence and map the genomes of thousands of individuals, with the help of a technology called HiSeq X, a high-throughput sequencer from Illumina. The HiSeq X produces 1.8 Tb of sequence data per run in 3 days and is specifically designed for whole-genome sequencing, which requires very high-throughput and simultaneous multiparallel sequencing.31 The target of whole-genome sequencing for $1000 is in sight, and the cost might be even lower with third-generation technologies.31 Currently, exome sequencing is already quite advanced in cardiovascular genetic studies32 and will gradually be followed by sequencing of the whole genome.
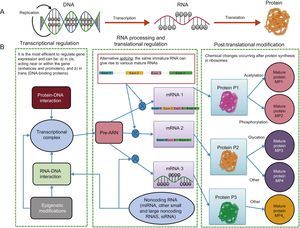
EPIGENOMICSDespite the considerable advances in genome sequencing, understanding of the genetic variants in DNA is insufficient to predict disease risk because there are other more dynamic regulatory elements, called the epigenome, that are also able to regulate the expression of DNA sequences. Accordingly, it is essential to study the genome and epigenome jointly to better understand the molecular bases of the cardiovascular risk phenotypes. The existence of epigenetic regulation helps to explain why the fundamental dogma of biology is not followed (Figure 4). In the classic process shown in panel A, DNA is transcribed to RNA and RNA is translated into protein, whereas panel B shows the more complex currently understood process, with a single DNA able to give rise to different proteins via the influence of distinct epigenetic regulatory processes that go beyond DNA mutations.13 Epigenomics is the study of the key functional elements regulating cellular gene expression. In contrast to the genome, which is the same in all somatic cells, the epigenome is specific for each cell type, increasing the complexity of sequencing studies and placing greater emphasis on the origin of the sample used for the analysis. There will be differences between epigenome analysis results if leukocytes or adipose tissue, for example, are analyzed, even when obtained from the same person.20 To analyze the epigenome of different cell types, various consortiums, brought together under the umbrella of the International Human Epigenome Consortium, have taken on the challenge of decoding the epigenome sequences of hundreds of healthy and unhealthy people, specific for each cell type, and are disseminating the results in various publications.33 Of the numerous types of epigenetic modifications, the most widely studied are methylations and regulations by noncoding RNA such as microRNA and other types of RNA. Histone modifications, another type of epigenetic regulation, are more complex and are still little studied in human epidemiological studies. The huge interest in improving our understanding of epigenetic regulators lies in their dynamicity. In contrast to SNPs in DNA, epigenetic marks can be modified and understanding of the factors involved in this modification might be crucial for the prevention and/or treatment of cardiovascular diseases.
Transcription and translation. A: According to the central dogma of biology, with a single gene giving rise to a single protein. B: The modern view, which includes various additional regulatory elements that allow the same gene to give rise to various proteins. miRNA, microRNA; MP, mature protein; mRNA, messenger RNA; Pre-RNA, precursor RNA; siRNA, silencing RNA.
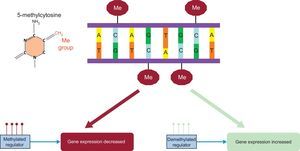
This type of epigenetic modification of DNA involves the enzymatic addition of a methyl group to the C-5 carbon of cytosine (Figure 5). Most 5-methylcytosines are present on cytosine-phosphate-guanine (CpG) dinucleotides. The DNA methylation is performed by DNA methyltransferases. These enzymes are classified into 2 groups: maintenance DNA methyltransferases (DNMT1), which maintain methylation patterns, and de novo DNA methyltransferases (DNMT3A and DNMT3B), which perform new methylations. Demethylases are in charge of the inverse process (ie, methyl group elimination) but these mechanisms are less well understood. Greater detail on all of these methylation and demethylation processes can be found in specific themed reviews.20,34,35 The CpG dinucleotides are not uniformly distributed throughout the genome but are concentrated in certain regions. Areas enriched in these dinucleotides (> 60%) are called CpG islands. These islands are usually concentrated between the promoter and the transcription start site. Methylation-dependent epigenetic regulation is complex but methylation of regulatory elements in genes, such as promoters, enhancers, isolators, and repressors, generally suppresses gene function (Figure 5).35 There are various ways to analyze the methylation of a specific gene or the genome as a whole. Accordingly, as discussed for whole-genome genotyping, in epigenome-wide association (EWA) studies of methylation, specific chips are used to analyze the most important regions of the genome. The initial 450K methylation chip of Illumina has recently been replaced with a chip with greater coverage, 850K, also produced by Illumina.36 To analyze associations, Manhattan plots are also used but, in these studies, the analyses evaluate the P values for the association between the methylation and the specific phenotype.37 Although various EWA studies have been published, the agreement among the results of these studies is still low. Considering other types of methylation, a review has analyzed the results of 31 articles focused on methylation and plasma lipids that included 8027 participants.38 Generally, there were no firm associations between general DNA methylation and plasma lipids. Regarding the methylation of specific genes, consistent results were found with ABCG1, CPT1A, TNNT1, MIR33B, SREBF1, and TNIP,38 which is why more in-depth study is required of these relationships and their modulators. Because dietary factors and smoking strongly influence DNA methylation,39 their effects need to be strongly considered in EWA studies.
Interest has also recently grown into hydroxymethylation regulation of DNA.40 This process is carried out by TET (ten-eleven translocation methylcytosine dioxygenase) enzymes, which recognize certain previously methylated cytosines and oxidize them, converting 5-methylcytosine to 5-hydroxymethylcytosine. Hydroxymethylated genes appear to be associated with increased transcription. Because this is a relatively new field, many more studies are required to determine the influence of hydroxymethylation on cardiovascular risk.
Regulation by Methylcytosine-phosphate-guanine–binding ProteinsThis mechanism involves proteins or protein complexes that specifically bind to methylated CpG regions and indirectly block the binding of transcription factors by limiting their access to regulatory elements.35 These proteins contain conserved methylated DNA-binding domains called methyl-binding domains (MBDs). The first protein identified was MeCP2, which was followed by other proteins, such as MBD1, MBD2, and MBD3, involved in transcriptional repression, the same as MeCP2. Other MBD proteins have distinct functions. This type of regulation is generally complex.35
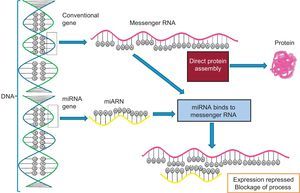
Regulation by Noncoding RNA. MicroRNA and OthersA large number of noncoding RNA transcripts have been identified. Their function was unknown for many years but progress is now being made in understanding their roles.41 These RNA do not encode proteins but play important regulatory roles in many processes. They are classified as short noncoding RNA (< 200 base pairs) and long noncoding RNA (> 200 base pairs).19 The shortest and best studied are microRNA (∼20-25 base pairs). microRNA are encoded in the genome, either in the introns within another coding gene or in the intergenic spaces. They are transcribed as a larger double-stranded primary transcript (pri-miR) by RNA polymerase II. Subsequently, the nuclear enzymes Drosha (also known as ribonuclease III) and Pasha convert this precursor into a double-stranded 70-nucleotide microRNA precursor (pre-miR), and a mechanism involving the protein exportin transports it to the cytoplasm. Finally, the enzyme Dicer processes this precursor into a double-stranded 22-nucleotide microRNA. This duplex is unrolled and 1 of the 2 strands is incorporated into the RISC (RNA-induced silencing complex), which comprises the protein argonaute, among others.42 The microRNA species incorporated in the RISC are able to bind to the 3’-UTR of target messenger RNA and block translation. The other microRNA strand is degraded. Figure 6 shows a schematic representation of the binding of microRNA to its target messenger RNA that blocks protein production and a comparison of the process without microRNA interference. Multiple microRNAs are involved in the regulation of the distinct cardiovascular phenotypes. These microRNAs can be analyzed in specific tissues and their expression related to certain phenotypic characteristics of tissues, such as, for example, the relationship between the expression of the microRNAs miR-1, miR-133, and miR-208 and the development of ventricular hypertrophy and heart failure. Additionally, microRNA can be found in the plasma circulation and analysis of their concentrations can indicate various diseases and processes. For example, the plasma levels of miR-1, miR-133a, miR-133b, and miR-499-5p are elevated after an acute myocardial infarction.42 This emerging research field, despite its considerable spread, requires better standardization of techniques and processes because it is subject to highly dynamic variation. After this standardization, multiple applications will no doubt be found for this fascinating world of epigenomic regulation.
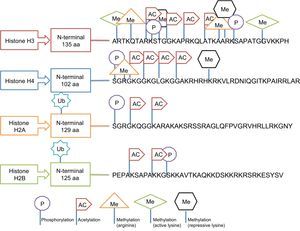
Regulation by Histone ModificationThis type of epigenetic regulation is more complex and less well-analyzed in human epidemiological studies. The main post-translational modifications that can take place in histones include acetylation, phosphorylation, methylation, and ubiquitination and influence the compaction of chromatin.20 Histones have a globular carboxy terminus and an amino terminal tail, which is where the modifications occur. However, these modifications do not occur in any site but respond to well-defined regulatory codes. Thus, for example, methylation occurs in lysine (abbreviated as K) and arginine (R) residues, acetylation in lysine residues, ubiquitination in lysine, and phosphorylation in serines (S) and threonines (T). The outcome is also affected by the type of histone being modified.35 This selective modification of histones is illustrated in Figure 7. Despite the difficulties, several studies have reported that HDAC3 (histone deacetylase 3) plays a critical role in endothelial function, whereas HDAC7 (histone deacetylase 7) is important for the function of vascular smooth muscle cells.35 In addition, histone enzymes tend to be more underexpressed than overexpressed in metabolic diseases.43 However, standardization is still required of the differences in their expression according to tissue and other complex regulators that also participate in the processes.35
GENOMIC AND EPIGENOMIC INTEGRATION. GENE-ENVIRONMENT INTERACTIONSAlthough the genomic and epigenomic concepts have been presented separately, the reality is that they considerably interact. For example, methylation depends on certain genetic polymorphisms that can be found in the sequence.44 Similarly, the binding of a microRNA to a target messenger RNA will differ if there is a specific base sequence or a genetic polymorphism.44 Thus, some of our interesting results can serve as an example. In our work, we analyzed a polymorphism at the binding site of a microRNA (in this case, miR410) in the lipoprotein lipase gene on plasma triglyceride concentrations and its subsequent modulation by the Mediterranean diet to determine stroke risk.45 All of these factors indicate the need for integrated studies, not only of -omics, but also of environmental factors such as diet and other life style variables, to better understand the molecular bases of cardiovascular risk and their regulation. To study gene-environment interactions, the proposed approach has been to explore the statistical interaction, instead of focusing on biological interactions (see a review46 to better understand the differences between them). However, the publication of an important biological, but not statistically significant, interaction between a GRS of cardiovascular risk and life style that determines the incidence of cardiovascular disease47 has helped to support the concept of a gene-environment interaction, understood as the existence of an environmental factor that can modify genetic susceptibility, and has led to new studies being planned from this perspective.
FUNDINGThis work has been supported by the Spanish Ministry of Health (Instituto de Salud Carlos III) and by the Ministry of Economy, Industry and Competitiveness (CIBER 06/03, CNIC-06/2007, RTIC G03/140, SAF2016-80532-R, AES_FIS_2016), and the TV3 Marathon Foundation (538/U/2016).
CONFLICTS OF INTERESTNone declared.