Cardiovascular diseases currently have a major social and economic impact, constituting one of the leading causes of mortality and morbidity. Personalized computational models of the heart are demonstrating their usefulness both to help understand the mechanisms underlying cardiac disease, and to optimize their treatment and predict the patient's response. Within this framework, the Spanish Research Network for Cardiac Computational Modelling (VHeart-SN) has been launched. The general objective of the VHeart-SN network is the development of an integrated, modular and multiscale multiphysical computational model of the heart. This general objective is addressed through the following specific objectives: a) to integrate the different numerical methods and models taking into account the specificity of patients; b) to assist in advancing knowledge of the mechanisms associated with cardiac and vascular diseases; and c) to support the application of different personalized therapies. This article presents the current state of cardiac computational modelling and different scientific works conducted by the members of the network to gain greater understanding of the characteristics and usefulness of these models.
Keywords
Cardiovascular pathologies have a major social and economic impact in Spain, and in the rest of the world, in terms of morbidity, mortality and cost for the health care system. The diagnostic and therapeutic assessment of patients still depends on empirical studies in which the results are compared statistically between large groups of patients with similar pathology. The choice of the optimum treatment is difficult and treatment efficacy is limited because each patient has a unique disease profile. Computational models of the heart (also called virtual heart, in silico heart, or digital twin) are emerging as a helpful tool by providing a framework to integrate various data from individual patients, which can be used to improve diagnosis and for optimize and plan personalized treatments.1
Biophysically based multiscale mathematical models of the heart provide a bridge between cellular models at smaller scales, and organ function at larger scales. For instance, in cardiac electrophysiology, experimental and clinical data are integrated by improved computational models of human action potential2 and its propagation in 2-dimensional (2D) and 3-dimensional (3D) cardiac structures. These models are being used to test and generate hypotheses that are difficult to address experimentally. Nondiseased cardiac models of human atria3,4 and ventricles5 have been built with a high degree of electro-anatomical detail. Models of the human torso are also currently available,6 allowing the reconstruction of the electrical field changes that generate the surface electrocardiogram.
To realistically simulate the complex multiphysics behavior of the heart, comprehensive computational models need to simulate electrophysiology, muscular contraction, and blood flow mechanics.7 In the last decade, advances in mathematical modelling and computational resources currently allow resolution of highly complex, tightly coupled, multiphysics models that can take into account feedback mechanisms at the biophysical level to perform reliable in silico studies, which are useful for patient risk stratification, treatment planning, or in silico clinical trials, among others.8,9
The increasing availability of in vivo cardiac images together with the rising trend toward personalized medicine has enabled the creation of patient-specific models. The cardiac anatomy of a particular human individual can be extracted from in vivo medical images (eg, computed tomography or magnetic resonance [MR]). Parameters estimated from population-based statistics or ex vivo experiments (fiber orientation, anisotropy, electrophysiological heterogeneity, among others) can also be incorporated into the models when patient-specific data are not available, ensuring that these parameters stay within the physiological range. The patient-specific model is then obtained by adjusting relevant parameters so that the results of the model simulation are in agreement with clinical recordings.
However, the road from the development of a patient-specific cardiac model to its clinical application is complex and requires a multidisciplinary team. Clinicians provide the anatomical-physiological data and prior knowledge to build the models, personalize and validate them. Professionals with different mathematical, engineering and physics backgrounds will formulate the equations and develop the software needed to solve them and run simulations that mimic heart multiphysics phenomena in healthy and pathological conditions. Recently, the Spanish Research Network for Cardiac Computational Modelling (VHeart-SN) was founded, including 9 well known Spanish universities and research centers with ample experience in different aspects of cardiac computer models. The main goal of the network is to develop an integrated, modular and multiscale multiphysics computational model of the heart and to increase collaborations with clinicians and medical companies, ultimately contributing to the translation of virtual heart models into clinical applications. This article summarizes the current state of cardiac computational modelling and different scientific works developed by the partners of the network.
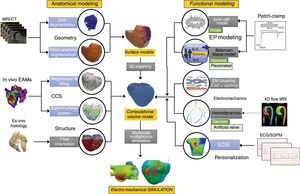
METHODSAnatomical modellingThere are a large number of available techniques to obtain a patient-specific 3D heart model from in vivo images acquired by MR or computed tomography10 (figure 1, geometry). Among them, it is worth mentioning those based on a-priori knowledge of the heart anatomy such as those based on statistical atlases,11 or more recent ones based on deep learning techniques.
Schematic representation of the pipeline to build a 3-dimensional (3D) cardiac computational model. This flowchart shows the main stages in the construction process of a 3D cardiac model aimed at biophysical simulation: 3D cardiac geometry generation and meshing (from magnetic resonance imaging [MRI] or computed tomography [CT]), cardiac conduction system (CCS) generation, myocardial structure generation, biophysical modelling (cardiac electrophysiology [EP] and electromechanics), and finally multiscale multiphysics simulation. Models can be personalized by means of patient electrocardiogram (ECG) or body surface potential maps (BSPM). Arrows show the flow from the clinical/biological data (ex vivo and in vivo) to the final computational model and electromechanical simulation. Blue boxes correspond to methods that allow us to obtain a patient-specific personalization of specific model properties, while gray boxes represent population-based or generic descriptions of other properties. ECGi, electrocardiographic imaging.
In addition to the 3D geometry, every cardiac computational model has to include other properties: cardiac fiber orientation, or pathologies affecting the myocardial structure (figure 1, cardiac conduction system and structure). Therefore, one has to use mathematical models to define the preferential direction in which the electrical excitation6,12 or mechanical deformation will propagate to its neighbors. The last critical anatomical component required in cardiac models is the cardiac conduction system. Some existing cardiac models have included the cardiac conduction system at a functional level (fast endocardial propagation), or have been based on ex vivo data.13 More recent advanced methods include the Purkinje network of a given patient by inverse estimation based on electro-anatomical map data.14
In addition to those anatomical components, it is possible to define regions where structural and functional remodelling is observed, such as scar or fibrotic tissue. These fibrotic lesions can be segmented from in vivo delayed enhancement MR,5 which provides accurate information about the location of ischemic injuries (infarct scar and the border zone).
Electrophysiological and mechanical modelsThe application of different experimental methods, such as patch-clamp, has allowed quantification of the biophysical properties of cardiac myocytes both at the cellular and tissue levels. This knowledge enabled the development of computational models of individual myocytes (figure 1, electrophysiological models). In general, they are based on the mathematical formalism of Hodgkin-Huxley,15 in which the cellular action potential is described by a system of equations that models the kinetics of individual ion channels, pumps and exchangers, as well as their electrical interactions. Several computational models of cellular cardiac electrical activity have been developed, including ventricular2 and atrial16 myocytes, and Purkinje fibers.17 Commonly used tissue propagation models are the bidomain model and the monodomain model,18 which are coupled to cellular models to form a multiscale model. These tissue models (1-dimensional [1D], 2D or 3D) can include the heterogeneous structure of cardiac tissue and heterogeneous expression at the cellular level (apical-basal, endocardial-epicardial and left-right ventricles), giving them a high level of realism.19
3D cardiac models aimed at electromechanical computational simulations must include electromechanical coupling, also known as excitation-contraction coupling. In this, electrical activation induces contraction of the cardiac tissue, which in turn influences the action potential through the stretch activated currents.20 Contraction is initiated by an increase in intracellular calcium, whose dynamics also has to be considered, to account for the appearance of several arrhythmias.21
Hemodynamics modelsThanks to computational fluid dynamics (CFD), we are able to mathematically obtain the behavior of the blood inside the heart. Heart models take into account large-scale motion and deformation, fluid-structure interaction of the valves, complex-flow induced dynamics inside the chambers and the transmission of the electrical impulse through the tissue, among other factors. We can solve all these coupled phenomena computationally using multiscale and multiphysics computational codes.8,22 With regard to model personalization, advanced diagnostic techniques, eg, 4-dimensional (4D) phase-contrast MR or 3D/4D ultrafast echocardiography have allowed us to obtain the complex blood flow inside the vessels and heart of the patient. Hence, combining these image technologies with CFD allows us to personalize the numerical simulation to obtain patient-specific computational models.23
Simpler computational models, known as 0D/lumped models, are also interesting computational tools to study hemodynamics in the whole cardiovascular system, which cannot be achieved with higher-dimensional models such as CFD and fluid-structure interaction. For example, a fetal circulation model, personalized with available Doppler ultrasound data were developed to quantify blood redistribution in cases of intrauterine growth restriction.24 Lumped models were also recently employed to study heart-brain interactions holistically, personalizing cardiovascular models with data from thousands of cases from the UK Biobank dataset, and finding new relations between neurological factors such as white-matter hyperintensity lesions and cardiac abnormalities such as atrial fibrillation (AF).25 In that line, it is also possible to create a reduced order model based on coupling 1D or 0D models with 3D models to study hemodynamics problems efficiently.26 1D models allow us to investigate physical mechanisms underlying changes in pressure and flow pulse waveforms that are produced by a cardiovascular disease27.
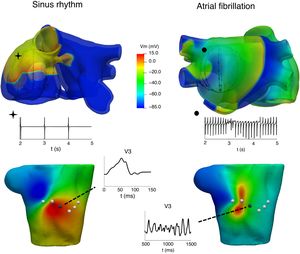
CLINICAL APPLICATIONSAtrial arrhythmiasIt is well known that atrial arrhythmias can be caused by various mechanisms, including single-circuit reentry, multiple-circuit reentry, rapid local ectopic activity, and rotors. Unravelling the mechanisms underlying atrial arrhythmias can have an important impact in tailoring treatment to individual patients or populations. One of the applications of computational models is in helping to understand the relationship between atrial activation patterns and the characteristics of electrograms recorded by catheters8,28,29 and in proposing improved electrogram biomarkers that can provide information about AF dynamics and the site of the AF drivers. Figure 2 shows the relationship between 2 atrial activations, in sinus rhythm (left) and AF (right), in electrograms recorded through an intracavitary catheter, body surface maps and P-wave at the V3 precordial lead.
At the top, atrial propagation produced by a sinus rhythm (left) and by an ectopic pacing from the coronary sinus (right) and their electrogram signals registered at 1 intracavitary electrode. At the bottom, body surface potential maps at 1 instant of the above simulations and P-wave registered at the V3 precordial lead.
Computer models also are a good platform to study interpatient electrophysiological differences as well as the contribution of patient-specific remodelling (ie, electric remodelling, fibrosis distribution) to AF initiation and maintenance, which can serve to guide treatment. Using 2D tissue models and 3D patient-specific models of the left and right atria, it has been shown that fibrosis and atrial wall thickness gradient can affect the stabilization of AF drivers.30
Additionally, to gain knowledge on AF initiation and perpetuation mechanisms, computer simulations could play a role in planning and predicting the effect of a treatment in the near future. AF ablation is nowadays the first-line treatment for the control of the rhythm of AF in selected patients, mostly patients with paroxysmal AF who prefer an interventional treatment.31 However, it is well known that the results of AF ablation depend on many important factors, such as the type of AF (paroxysmal, persistent, or permanent), age, comorbidities, atrium size, the use of antiarrhythmic drugs, and the number and time of repeated ablation procedures. The major challenge in AF ablation procedures is correct patient selection and identification of the adequate size of the generated lesions to avoid unnecessary damage to the atrium. The efficacy of AF ablation may be improved with an optimized patient-specific ablation strategy predicted by a computer model prior to an ablation procedure. For instance, an in silico study recently demonstrated the thermal impact of balloon occlusion of the coronary sinus during mitral isthmus radiofrequency ablation.32
As proof of concept that computational models can guide ablation procedures, Boyle et al.1 carried out a prospective study using 10 patients with persistent AF and atrial fibrosis for targeted optimum ablation. The authors used personalized computational models of the atria reconstructed from each individual's late gadolinium enhancement MR imaging scans to identify the sites that were maintaining the AF or that could be potential sources of AF in the future. They claim that the use of this methodology increases the efficacy of the procedure.
The location of ectopic foci during focal atrial tachycardia is difficult to determine by noninvasive techniques. Methods based on either standard 12-lead electrocardiograms or body surface potential maps have not shed sufficient light on how to accurately correlate ectopic focus locations with the distribution of potential and P-wave characteristics on the torso surface. This limitation results in longer intraoperative invasive mapping procedures, which could lead to suboptimal ablation of focal triggers. Recently, a highly detailed multiscale atrial-torso model has been used to develop a machine learning methodology to classify body surface potential maps into groups associated with ectopic atrial foci sites.6,33
Computational modelling has also made additional contributions to the characterization of autonomic effects on atrial electrical activity and the establishment of actions leading to atrial arrhythmia termination or perpetuation, with recent interest in modelling the intrinsic cardiac autonomic nervous system to assess ganglionated plexi ablation in combination with pulmonary vein isolation.34
Ventricular tachycardias and cardiac resynchronisation therapy intervention planningElectrophysiological ventricular models have helped to shed light on the mechanisms underlying ventricular repolarization phenomena35,36 and to improve the definition of arrhythmic risk markers quantified from electrogram and electrocardiogram signals. As an example, low-frequency oscillations of ventricular repolarization, recently proposed to guide prophylactic implantable cardioverted-defibrillator implantation,37 has been investigated through computational modelling and simulation to uncover its cellular bases, establish a relationship with arrhythmic risk, and link it to other arrhythmogenic phenomena.38,39 Another recent patient-specific electrophysiological model, called in silico pace-mapping, predicted the right outflow tract origin in idiopathic ventricular arrhythmias in 10/11 cases.40 A patient-specific model has also been used to investigate the mechanism of microreentry that leads to the generation of ectopic beats near infarct areas.41,42 The simulation results explain how ectopic beats emerge from microreentries that are sustained by the heterogeneous structure of the infarct regions.
Cardiac resynchronisation therapies, also called biventricular pacing, is the most widespread treatment for this kind of cardiac malfunction. A comprehensive fluid-electro-mechanical model of the heart, involving blood dynamics inside the cardiac cavities, becomes an invaluable predictive simulation tool to analyze cardiac resynchronisation therapy at different stages and by different stakeholders, such as medical technology companies to improve design and deployment, or for practitioners to improve treatment by adapting it to a given patient.
The cardiac computational model implemented within the Alya code7–9 comprises the main mechanisms for the function of the cardiac muscle including the electrophysiology, the cardiac muscle and blood flow mechanics, the excitation-contraction-coupling and the mechano-electric feedback to study the cardiac muscle in a holistic manner. Due to this model, it is possible to simulate the cardiac resynchronisation therapy performed using a traditional pacemaker (figure 3, above) and a transcatheter intraventricular pacemaker (figure 3, below). In particular, in the second case, fluid mechanics inside the ventricles are analyzed through the vortices generated by the pacemaker.
Transcatheter intraventricular pacemaker. Above, cardiac wall stresses evolution during pacing. The tissue is colored by the VonMises stress. Below, blood mechanics evolution inside the ventricles. The tissue is colored by the electrical activity propagation. Fluid mechanics are analyzed through the vortices produced.
Another application where numerical models have been very helpful in improving the technology is the design of pacemakers and defibrillators.43 State of the art numerical studies have shown that the fiber orientation and membrane kinetics (electroporation) are crucial in determining the outcome of defibrillation.44 The ultimate goal of an optimized personal electrical device to support the failing heart has yet to be achieved. Despite improvements in this direction, we still need to push further the computational models to accomplish this objective. The major insights uncovered by computer models are that realistic heart geometry and disease remodelling are important.43
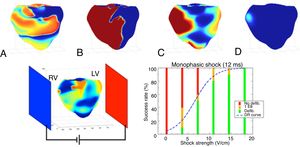
Realistic 3D numerical simulations of the defibrillation problem were conducted and analyzed in a ventricular geometry. Upon increase of the shock strength, the defibrillation success rate increases and follows the classic logistic curve (dose-response curve), as shown in figure 4. The simulation data were in good agreement with the clinical data. In subsequent studies, we have used numerical simulations to help in the design of the ideal waveform45 and electrode placement.46 Much more research is still needed to find the optimum strategy for defibrillation.
Above, 4 consecutive snapshots taken during a successful defibrillation shock of 12ms duration. A: electrical activity before the shock. B: strong polarized activity during the shock. C: activity 50ms after the end of the shock. D: no activity 100ms after the end of the shock. Below, setup and plot showing the defibrillation success rate as a function of electrical shock strength as obtained through 50 numerical simulations. The logistic curve (dose-response curve [DR] curve) represents the best fit of the data. LV, left ventricle; RV, right ventricle.
Heart hemodynamics can also contribute to a better understanding of blood flow patterns in complex cardiac chambers such as the left atria and optimize medical devices such as left atrial appendage occluders (LAAO). In patients with contraindications to oral anticoagulants, LAAO devices are implanted to prevent blood flow entering the left atrial appendage, where 99% of the thrombi related to AF are formed. Suboptimal LAAO implantation can generate thrombi outside the device. A web-based 3D interactive virtual implantation platform,47 called VIDAA, was created to select the most appropriate LAAO configurations for a given patient-specific left atrial appendage morphology, predicting the region of peridevice thrombus formation in some cases.48 Related to cardiovascular hemodynamics, the US Food and Drug Administration approved the clinical tool HeartFlow FFRCT Analysis to assess fractional flow reserve. HeartFlow's FFRCT software was one of the first CFD codes to be used in routine clinical practice, opening the way to advanced computational codes such as CaaS 3D-QCA vFFR (Pie Medical Imaging, The Netherlands). The 3D-QCA vFFR workflow builds a 3D reconstruction of the coronary artery by combining orthogonal angiographic projections and assesses pressure drop, resulting in a virtual fractional flow reserve value using a CFD code.27
DISCUSSIONThe technological progress during the last few years, including advanced high-computing infrastructures, open-source software and open-access medical databases, have brought biophysical models closer to clinical translation. They are currently being used in academia and industry for a better understanding of the physiology and for the optimization of medical devices and therapies. However, modelling-based tools are rarely employed in other clinical decisions such as diagnosis and treatment recommendations.
There are several reasons for not having a more established use of biophysical models in routine clinical practice. The necessary large process of clinical validation imposes robustness and time requirements that are not simple to achieve. Although several clinical research studies based on biophysical models have shown their potential, they still need to demonstrate their added value in randomized clinical trials. Even if difficult, setting up multidisciplinary teams, jointly defining projects from the very beginning is essential to bring biophysical models closer to routine clinical practice. Biophysical models require prior knowledge by clinicians to obtain realistic simulations mimicking the complex behaviour of the heart. Moreover, computational tools are usually data-hungry but not all types of clinical data is useful: data need to be appropriate to the question that the model is designed to answer, and should ideally be acquired prospectively. Furthermore, data should be fully curated and annotated, which is often a tedious and time-consuming process. Biomedical engineers need to be embedded in clinical units with a modern information technology infrastructure to provide the required technological skills and tools to perform data-related tasks such as harmonization, anonymization, and standardization. Moreover, large databases, with an appropriate balance of controls and pathological cases with events, require multisite studies, with clinical centers adopting data sharing philosophies, always fulfilling data protection laws. Finally, implementation efforts are required to progress from the development of tools in an academic environment to regular use in clinical workflows, creating clinician-friendly and application-specific visualization interfaces to search, integrate and explore patient-specific multimodal data, population-based statistics, simulation results, and clinical knowledge. Software environments such as the Rocket ecosystem49 are currently being developed to tackle these challenges.
The future of computational tools in cardiology is bright; we are at the beginning of a paradigm shift in medicine in which advanced biophysical models will play a significant role in cardiology pathways. During the next few years, biophysical models will significantly improve due to the strong involvement of clinicians in their design and development, defining the most relevant questions to be answered, and incorporating high-quality patient-specific data to validate the added value of the simulations. Therefore, clinicians must not fear computational tools but should fully embrace them; biophysical models and artificial intelligence algorithms will not replace clinicians but will support them, allowing a focus on more interesting human-related tasks such as reasoning and patient-physician communication.
In Spain, there are excellent research teams with worldwide experts in their respective medical and engineering fields. The VHeart-SN initiative helped to integrate the national research groups working on cardiac modelling, in the spirit of sharing knowledge and resources. The next step should be to fully integrate medical teams in the initiative and form multidisciplinary units to ease the clinical translation of computational tools to improve clinical decisions for our patients and foster innovation through connection with large health care companies and the creation of start-ups.
FUNDINGThis work was partially supported by: Acciones de Dinamización Redes de Excelencia 2016, Plan Estatal de Investigación Científica y Técnica y de Innovación, Ministerio de Economía y Competitividad (DPI2016-81873-REDT) and CompBioMed2, Grant agreement ID: 823712. The authors also thank the support of the European Research Council (ERC-StG 638284) and the Spanish Goverment through the following programmes: Retos I+D (TIN2014-59932-JIN, RTI2018-093416-B-I00, SAF2017-88019-C3-3R, RTI2018-101193-B-I00, DPI2016-75799-R and PID2019-105674RB-I00); María de Maeztu (MDM-2015-0502); and Severo Ochoa (SEV-2017-0718 and CEX2018-000797-S).
CONFLICTS OF INTERESTThe authors have nothing to disclose.


![Schematic representation of the pipeline to build a 3-dimensional (3D) cardiac computational model. This flowchart shows the main stages in the construction process of a 3D cardiac model aimed at biophysical simulation: 3D cardiac geometry generation and meshing (from magnetic resonance imaging [MRI] or computed tomography [CT]), cardiac conduction system (CCS) generation, myocardial structure generation, biophysical modelling (cardiac electrophysiology [EP] and electromechanics), and finally multiscale multiphysics simulation. Models can be personalized by means of patient electrocardiogram (ECG) or body surface potential maps (BSPM). Arrows show the flow from the clinical/biological data (ex vivo and in vivo) to the final computational model and electromechanical simulation. Blue boxes correspond to methods that allow us to obtain a patient-specific personalization of specific model properties, while gray boxes represent population-based or generic descriptions of other properties. ECGi, electrocardiographic imaging. Schematic representation of the pipeline to build a 3-dimensional (3D) cardiac computational model. This flowchart shows the main stages in the construction process of a 3D cardiac model aimed at biophysical simulation: 3D cardiac geometry generation and meshing (from magnetic resonance imaging [MRI] or computed tomography [CT]), cardiac conduction system (CCS) generation, myocardial structure generation, biophysical modelling (cardiac electrophysiology [EP] and electromechanics), and finally multiscale multiphysics simulation. Models can be personalized by means of patient electrocardiogram (ECG) or body surface potential maps (BSPM). Arrows show the flow from the clinical/biological data (ex vivo and in vivo) to the final computational model and electromechanical simulation. Blue boxes correspond to methods that allow us to obtain a patient-specific personalization of specific model properties, while gray boxes represent population-based or generic descriptions of other properties. ECGi, electrocardiographic imaging.](https://static.elsevier.es/multimedia/18855857/0000007400000001/v1_202012200625/S188558572030270X/v1_202012200625/en/main.assets/thumbnail/gr1.jpeg?xkr=eyJpdiI6Ik1UcE1KT3VWK2VEMDFFcnNpU3cwa0E9PSIsInZhbHVlIjoiUThtWFd0OURpSm1lMVVXaElrcUthWjVZRmZveUhnTVJNM3ljT3NhcWh5WT0iLCJtYWMiOiI3N2ZkNTIwODM0MzM3NWEyM2YyMmJiMGExNjg5NTZlNTY1YjI2ODBiMjNiODQxYTU1ZTczZWQyYTE0Y2NiOGIyIiwidGFnIjoiIn0=)


![Above, 4 consecutive snapshots taken during a successful defibrillation shock of 12ms duration. A: electrical activity before the shock. B: strong polarized activity during the shock. C: activity 50ms after the end of the shock. D: no activity 100ms after the end of the shock. Below, setup and plot showing the defibrillation success rate as a function of electrical shock strength as obtained through 50 numerical simulations. The logistic curve (dose-response curve [DR] curve) represents the best fit of the data. LV, left ventricle; RV, right ventricle. Above, 4 consecutive snapshots taken during a successful defibrillation shock of 12ms duration. A: electrical activity before the shock. B: strong polarized activity during the shock. C: activity 50ms after the end of the shock. D: no activity 100ms after the end of the shock. Below, setup and plot showing the defibrillation success rate as a function of electrical shock strength as obtained through 50 numerical simulations. The logistic curve (dose-response curve [DR] curve) represents the best fit of the data. LV, left ventricle; RV, right ventricle.](https://static.elsevier.es/multimedia/18855857/0000007400000001/v1_202012200625/S188558572030270X/v1_202012200625/en/main.assets/thumbnail/gr4.jpeg?xkr=eyJpdiI6IlF3cHB1ZjlPNDRUb2cwbDhkMEliaVE9PSIsInZhbHVlIjoiQ2F2NUNpcnhrMHEvV1ZxR29NWUhqNzA3dVI1RW1tb3VLN2VuNE1idFczND0iLCJtYWMiOiIyOGE5OGY1ZGE1NjdjODM0NTVhOTZkZGI0MjkzODZiZjBmNTA5ZjQ5YjhlYTFlNzc1YmFmNjMwMzNmYWQxYTdhIiwidGFnIjoiIn0=)