The objectives of this study were to analyze the association between two genetic variants (rs2200733 and rs7193343) in a Spanish population and the risk of developing atrial fibrillation, and to carry out a systematic review and meta-analysis of these associations.
MethodsWe performed a case-control study involving 257 case patients with atrial fibrillation and 379 controls. The case patients were individuals who had donated samples to the Spanish National DNA Bank; the controls were participating in a population-based cross-sectional study. Genotyping was carried out using a TaqMan assay. We conducted a systematic literature search in which 2 independent reviewers extracted the necessary information. The study involved a meta-analysis, a heterogeneity analysis, and a meta-regression analysis to identify the variables that explain the heterogeneity across studies.
ResultsIn our population, the presence of atrial fibrillation was found to be associated with rs2200733 (odds ratio = 1.87; 95% confidence interval, 1.30-2.70), but not with rs7193343 (odds ratio = 1.18; 95% confidence interval, 0.80-1.73). In the meta-analysis, we observed an association between atrial fibrillation and both variants: odds ratio = 1.71 (95% confidence interval, 1.54-1.90) for rs2200733 and odds ratio = 1.18 (95% confidence interval, 1.11-1.25) for rs7193343. We observed heterogeneity among the studies dealing with the association between rs2200733 and atrial fibrillation, partially related to the study design, and the strength of association was greater in case-control studies (odds ratio = 1.83) than in cohort studies (odds ratio = 1.41).
ConclusionsVariants rs2200733 and rs7193343 are associated with a higher risk of atrial fibrillation. Case-control studies tend to overestimate the strength of association between these genetic variants and atrial fibrillation.
Keywords
Atrial fibrillation (AF) is the arrhythmia most frequently encountered in clinical practice1,2 and is associated with higher risk of mortality and incidence of cardiovascular disease.3 Among the factors that increase the risk of developing AF are age, hypertension, cardiomyopathies, and valve disease. Atrial fibrillation occurring in patients < 60 years of age with no clinical or echocardiographic evidence of cardiovascular disease is referred to as lone AF, and its prognosis is good.
There is substantial familial aggregation in the development of AF, indicating the relevance of genetic factors.4 Familial forms, although uncommon, have been reported, and mutations have been identified on different genes, generally related to ion channels.4 However, in the majority of the cases of AF found in the general population, individual susceptibility is related to multiple genes, genetic variants, and environmental factors. In recent years, genome-wide association studies have identified common genetic variants associated with a higher risk of AF.5,6 The 2 loci that most consistently have been associated with AF are found in the 4q25 region, in linkage disequilibrium with the PITX2 gene, where the single nucleotide polymorphism rs2200733 is of particular interest, and in the 16q22 region, associated with the ZFHX3 gene, where the single nucleotide polymorphism rs7193343 is located. Among the limitations of studies of this type are the possible variations between different publications. Some meta-analyses have analyzed the available evidence and the existence of heterogeneity among studies, but have failed to analyze the causes.7,8
The objectives of this study were as follows: a) to analyze the association between the abovementioned genetic variants, rs2200733 and rs7193343, and the probability of AF in a Spanish population, and b) to carry out a systematic review and meta-analysis of the association between these two genetic variants and the risk of AF, determining whether there was heterogeneity among the studies and, if so, its possible causes.
METHODSAnalysis of the Association Between Variants rs2200733 and rs7193343 and Atrial Fibrillation in a Spanish PopulationStudy DesignWe designed a case-control study in which the cases corresponded to patients with AF who had donated biological samples to the Spanish National DNA bank and from 2 hospitals participating in the project. One of the Spanish National DNA bank hubs collected samples from patients with cardiovascular disease, among them, AF. The patients from the Spanish National DNA bank were recruited in 4 hospitals between February 2007 and December 2009. In addition, we included patients with AF from 2 hospitals that recruited these individuals during 2011. The patients with AF ranged in age from 18 years to 70 years at the time of diagnosis, were Caucasian, and had AF documented in at least 1 electrocardiographic recording. Lone AF was defined as that found in patients younger than 60 years with no clinical or echocardiographic evidence of cardiovascular disease. Recruitment of these patients was performed prospectively, including all the cases of AF diagnosed during the study period, and retrospectively, in which case we contacted only those patients who had been diagnosed as having lone AF to ask them to participate in the study.
The controls were selected randomly from a population-based cross-sectional study carried out in Spain between 2000 and 2001 that included Caucasian individuals ranging in age from 19 years to 75 years.9 The presence of AF was ruled out on the basis of an absence of suspicious symptoms and a normal electrocardiogram.
All the case patients and controls signed an informed consent form prior to their inclusion in the study, which received the approval of the Clinical Research Ethics Committee of Hospital Universitario Sant Joan in Reus, a city in Catalonia, Spain, and the Scientific Committee of the Spanish National DNA Bank.
GenotypingDeoxyribonucleic acid was isolated from peripheral blood mononuclear cells using a standardized column method described elsewhere.10 The 2 genetic variants were genotyped using Taqman allelic discrimination assays (Life Technologies Ltd.; Paisley, United Kingdom) on the ABI 7900HT Real-Time PCR System (Life Technologies Ltd.).
Other VariablesInformation was collected on the history of hypertension and tobacco use. Hypertension was defined as a systolic blood pressure ≥ 140mmHg, a diastolic blood pressure ≥ 90mmHg, or use of antihypertensive medication. We considered a smoker to be any individual who had smoked, on average, ≥ 1 cigarette/day during the preceding week or any ex-smoker who had been abstinent < 1 year. The weight and height of the participants were recorded and the body mass index calculated (weight in kilograms divided by height in meters squared).
We had access to the following echocardiographic data for a subgroup of AF patients: left ventricular end-diastolic and end-systolic diameters, interventricular septal thickness and left ventricular posterior wall thickness, and left atrial anteroposterior diameter. The echocardiographic studies were performed and interpreted in the hospitals in which the AF case patients had been recruited in accordance with standardized international protocols.
Statistical AnalysisQuality control of the genotype data included the analysis of Hardy-Weinberg equilibrium. The continuous variables were tested for normality. For between-group comparisons, the chi-square test was used for categorical variables and Student t test or analysis of variance was employed for continuous variables. Logistic regression was utilized for the multivariate analysis. The association was considered to be statistically significant when P < .05.
Systematic Review and Meta-analysisReview of the LiteratureUsing the PubMed database, we carried out a search for articles on the subject of interest published up to 31 March 2013. The search strategy was based on the following terms: (rs2200733 or rs7193343 or 4q25 or 16q22 or PITX2 or ZFHX3) and (“Atrial fibrillation” or “supraventricular arrhythmias”) and (“genetics” or “polymorphism”). We also analyzed reviews on “atrial fibrillation” and “genetics”, and the bibliography of the articles ultimately selected in order to identify any article that had not been identified in the original search.
In a first step, 2 independent reviewers (AF and RE) analyzed the title and abstract of the articles identified and selected the candidates to review the entire article. The same reviewers, independently, extracted the information necessary for the meta-analysis and for the evaluation of the quality of the evidence presented. In case of disagreement, the 2 reviewers reached a consensus decision.
In the articles that included more than one population or study, the results of each of the studies included were considered individually. In the case of studies that were dealt with in several publications, we considered only the results of the article that included the largest number of participants.
Qualitative Evaluation of the Available EvidenceThe STREGA recommendations were followed for the evaluation of the quality and transparency of the information provided in the selected studies.11 These guidelines define 22 items that should be described or discussed in the different sections of the manuscripts. The two reviewers assigned a 1 or a 0 depending on the presence or absence of an adequate description of each of these items. The differences of opinion were discussed and resolved by consensus. The higher the value of this indicator, the higher the quality of the information presented in the article.
Quantitative Evaluation of the Associations of Interest: Meta-analysisFrom each of the selected studies, we obtained the following data: study design (case-control, cohort), type of AF (all types; lone; recurrence following ablation), number of case patients and controls included, estimate of the risk associated with each genetic variant and the 95% confidence interval (95%CI), whether or not the risk estimate was adjusted for confounding variables, frequency of the risk allele in the control population, ethnic origin, age, and the proportion of women and of hypertensive individuals.
We conducted a meta-analysis of the previously selected studies and also included the data from our study using the meta.DSL function of the R rmeta package, applying the random effects method of DerSimonian and Laird.12 In addition, we analyzed sensitivity by repeating the meta-analysis and eliminating a study each time to determine whether any of those included exerted a powerful influence on the strength of the association observed. The analyses were stratified for the evaluation of total and lone AF.
Analysis of the Heterogeneity and of Its Causes: Meta-regression AnalysisThe presence of heterogeneity was analyzed using the quantity I2, which describes the percentage of the variation across studies that is due to heterogeneity rather than chance.13 To analyze its causes, we performed a meta-regression analysis using a mixed effects model, and the factors that could explain the heterogeneity between studies were examined. The potential factors analyzed were study design (case-control vs cohort), number of case patients included (n ≥ 500 vs n < 500), adjustment for confounding variables, ethnic origin, and STREGA score (median < 18 vs ≥ 18). When any variable that could explain the heterogeneity was identified, the meta-analysis was repeated with stratification for that variable.
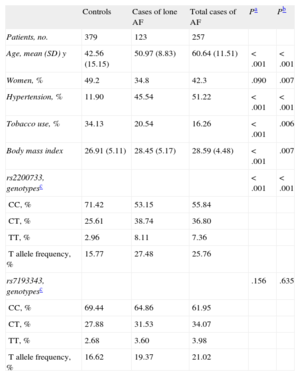
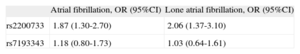
RESULTSAnalysis of the Association Between Variants rs2200733 and rs7193343 and Atrial Fibrillation in a Spanish PopulationThe sociodemographic characteristics, risk factor history, and genotype and allele frequencies of the genetic variants analyzed in the group of patients with AF (total and lone) and the control group are presented in Table 1. Table 2 shows the findings concerning the association between rs2200733 and rs7193343 and the presence of AF (total and lone); a statistically significant association was observed between rs2200733 and AF, but not between rs7193343 and AF. We analyzed a subsample of case patients (n = 150) and controls (n = 150), matched for age and sex, and the results were similar to those observed in the total sample.
Sociodemographic Characteristics, Risk Factor History, and Genotype and Allele Frequencies of the Genetic Variants Analyzed in the Group of Patients With Atrial Fibrillation and the Control Group
| Controls | Cases of lone AF | Total cases of AF | Pa | Pb | |
| Patients, no. | 379 | 123 | 257 | ||
| Age, mean (SD) y | 42.56 (15.15) | 50.97 (8.83) | 60.64 (11.51) | < .001 | < .001 |
| Women, % | 49.2 | 34.8 | 42.3 | .090 | .007 |
| Hypertension, % | 11.90 | 45.54 | 51.22 | < .001 | < .001 |
| Tobacco use, % | 34.13 | 20.54 | 16.26 | < .001 | .006 |
| Body mass index | 26.91 (5.11) | 28.45 (5.17) | 28.59 (4.48) | < .001 | .007 |
| rs2200733, genotypesc | < .001 | < .001 | |||
| CC, % | 71.42 | 53.15 | 55.84 | ||
| CT, % | 25.61 | 38.74 | 36.80 | ||
| TT, % | 2.96 | 8.11 | 7.36 | ||
| T allele frequency, % | 15.77 | 27.48 | 25.76 | ||
| rs7193343, genotypesc | .156 | .635 | |||
| CC, % | 69.44 | 64.86 | 61.95 | ||
| CT, % | 27.88 | 31.53 | 34.07 | ||
| TT, % | 2.68 | 3.60 | 3.98 | ||
| T allele frequency, % | 16.62 | 19.37 | 21.02 |
AF, atrial fibrillation; C, cytosine; T, thymine.
Unless otherwise indicated, data are expressed as mean (standard deviation).
Probability of Atrial Fibrillation Corresponding to the Different Genetic Variants Analyzed Defining an Additive Model
| Atrial fibrillation, OR (95%CI) | Lone atrial fibrillation, OR (95%CI) | |
| rs2200733 | 1.87 (1.30-2.70) | 2.06 (1.37-3.10) |
| rs7193343 | 1.18 (0.80-1.73) | 1.03 (0.64-1.61) |
95%CI, 95% confidence interval; OR, odds ratio.
Increase in the likelihood of atrial fibrillation due to the presence of each risk allele (T).
Results adjusted for age, sex, body mass index, history of hypertension, and tobacco use.
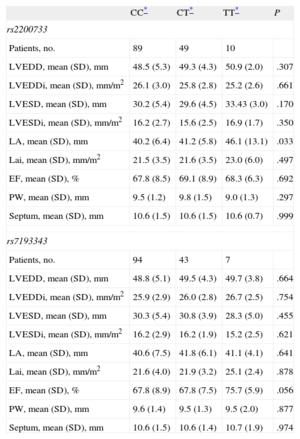
In the subgroup of patients with AF and echocardiographic data for left atrium (n = 148), we found an association between rs2200733 and the anteroposterior diameter, demonstrating that the AF risk allele was also associated with an enlarged left atrium (Table 3). This association lost statistical significance when the atrial diameter was indexed to body surface area. We observed no association between rs7193343 and the echocardiographic variables recorded.
Differences in the Echocardiographic Characteristics of the Case Patients With Atrial Fibrillation According to Genotype
| CC* | CT* | TT* | P | |
| rs2200733 | ||||
| Patients, no. | 89 | 49 | 10 | |
| LVEDD, mean (SD), mm | 48.5 (5.3) | 49.3 (4.3) | 50.9 (2.0) | .307 |
| LVEDDi, mean (SD), mm/m2 | 26.1 (3.0) | 25.8 (2.8) | 25.2 (2.6) | .661 |
| LVESD, mean (SD), mm | 30.2 (5.4) | 29.6 (4.5) | 33.43 (3.0) | .170 |
| LVESDi, mean (SD), mm/m2 | 16.2 (2.7) | 15.6 (2.5) | 16.9 (1.7) | .350 |
| LA, mean (SD), mm | 40.2 (6.4) | 41.2 (5.8) | 46.1 (13.1) | .033 |
| Lai, mean (SD), mm/m2 | 21.5 (3.5) | 21.6 (3.5) | 23.0 (6.0) | .497 |
| EF, mean (SD), % | 67.8 (8.5) | 69.1 (8.9) | 68.3 (6.3) | .692 |
| PW, mean (SD), mm | 9.5 (1.2) | 9.8 (1.5) | 9.0 (1.3) | .297 |
| Septum, mean (SD), mm | 10.6 (1.5) | 10.6 (1.5) | 10.6 (0.7) | .999 |
| rs7193343 | ||||
| Patients, no. | 94 | 43 | 7 | |
| LVEDD, mean (SD), mm | 48.8 (5.1) | 49.5 (4.3) | 49.7 (3.8) | .664 |
| LVEDDi, mean (SD), mm/m2 | 25.9 (2.9) | 26.0 (2.8) | 26.7 (2.5) | .754 |
| LVESD, mean (SD), mm | 30.3 (5.4) | 30.8 (3.9) | 28.3 (5.0) | .455 |
| LVESDi, mean (SD), mm/m2 | 16.2 (2.9) | 16.2 (1.9) | 15.2 (2.5) | .621 |
| LA, mean (SD), mm | 40.6 (7.5) | 41.8 (6.1) | 41.1 (4.1) | .641 |
| Lai, mean (SD), mm/m2 | 21.6 (4.0) | 21.9 (3.2) | 25.1 (2.4) | .878 |
| EF, mean (SD), % | 67.8 (8.9) | 67.8 (7.5) | 75.7 (5.9) | .056 |
| PW, mean (SD), mm | 9.6 (1.4) | 9.5 (1.3) | 9.5 (2.0) | .877 |
| Septum, mean (SD), mm | 10.6 (1.5) | 10.6 (1.4) | 10.7 (1.9) | .974 |
C, cytosine; EF, ejection fraction; LA, left atrial anteroposterior diameter; LAi, left atrial anteroposterior diameter indexed to body surface area; LVEDD, left ventricular end-diastolic diameter; LVEDDi, left ventricular end-diastolic diameter indexed to body surface area; LVESD, left ventricular end-systolic diameter; LVESDi, left ventricular end-systolic diameter indexed to body surface area; PW, posterior wall thickness; SD, standard deviation; Septum, septal thickness; T, thymine.
Unless otherwise indicated, data are expressed as mean (standard deviation).
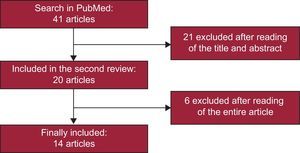
With the literature search criteria utilized, 41 articles were selected in a first stage, and were ultimately reduced to 14. A flow chart of the result of the literature search and article selection performed for this study is shown in Figure 1. Of the 14 studies selected, 10 analyzed the association between AF risk and rs220073314–23 and 4 analyzed the AF risk associated with rs7193343.6,24–26 These 14 articles included a total of 27 different studies, 19 that analyzed rs2200733 and 8 that focused on rs7193343; 22 were case-control studies and 5 were cohort studies.
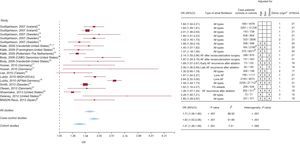
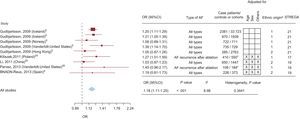
The scores resulting from the qualitative evaluation of the studies based on the STREGA guidelines ranged between 15 points and 21 points; those corresponding to each study are presented in Figures 2 and 3.
Graphical display of the results of the meta-analysis of studies on the association between rs2200733 and atrial fibrillation (forest plot), including the strength of association observed in individual studies, sample size, adjustment variables utilized, and the results of the evaluation according to the STREGA guidelines. The figure also shows the results of the heterogeneity analysis (I2), the estimator of the combined association in all the studies, and stratification according to study design (case-control or cohort). 95%CI, 95% confidence interval; AF, atrial fibrillation; BNADN: Banco Nacional de ADN (Spanish National DNA Bank); OR, odds ratio.
aAge < 60 years.
bResults of the study presented in this article.
c1, European; 2, Asian.
dCohort study.
Graphical display of the results of the meta-analysis of studies on the association between rs7193343 and atrial fibrillation (forest plot), including the strength of the association observed in individual studies, sample size, adjustment variables utilized, and the results of the evaluation according to the STREGA guidelines. The figure also shows the results of the heterogeneity analysis (I2) and the estimator of the combined association in all the studies. 95%CI, 95% confidence interval; AF, atrial fibrillation; BNADN: Banco Nacional de ADN (Spanish National DNA Bank); OR, odds ratio
aResults of the study presented in this article.
b1, European; 2, Asian.
cCohort study.
Figure 2 shows that the results of the meta-analysis demonstrated a statistically significant association between rs2200733 and AF (odds ratio [OR] = 1.71; 95%CI, 1.54-1.90). These findings were corroborated by the sensitivity analysis in which studies were eliminated one by one (OR between 1.62 and 1.75). For the analysis of lone AF, we included 6 studies, and the strength of the association was greater (OR = 2.16; 95%CI, 1.85-2.51). In the sensitivity analysis, as studies were eliminated one at a time, the OR ranged from 2.03 to 2.25.
We detected heterogeneity in the meta-analysis of the single nucleotide polymorphism rs220733 (total AF, I2 = 88.52; lone AF, I2 = 13.59). Meta-regression analysis indicated that one of the variables that partially explained this heterogeneity was the type of study design: case-control or cohort. When the meta-analysis was stratified for study design, we observed that there continued to be heterogeneity across the case-control studies (I2 = 51.69), that the strength of the association was greater than that of the cohort studies (OR = 1.83; 95%CI, 1.63-2.06 vs OR = 1.41; 95%CI, 1.26-1.59), and, moreover, that there was no heterogeneity among the latter (Figure 2).
There was a statistically significant association between rs7193343 and total AF (OR = 1.18; 95%CI, 1.11-1.25) (Figure 3). When studies were eliminated one by one in the sensitivity analysis, the OR ranged between 1.17 and 1.20. We observed no heterogeneity among studies (I2 = 8.98; P = .34). When we analyzed lone AF, only 2 studies were included and the strength of association was similar (OR = 1.11; 95%CI, 0.89-1.39)
We also estimated the population risk attributable to these 2 genetic variants using the OR from the meta-analysis and the prevalence of the genotypes of our population by applying the following formula: population attributable risk = prevalence of the risk genotype × [(OR – 1) / OR]. The population risk attributable to polymorphism rs2200733 was 13.77% and that of rs7193343, 4.99%.
DISCUSSIONIn this study, we observed an association between rs2200733 and risk of AF in a Spanish population. However, the association between rs7193343 and the presence of AF did not reach statistical significance. A systematic review and a meta-analysis of previous studies demonstrates an association between the 2 genetic variants analyzed and the risk of AF, but the strength of the association is greater in the case of rs2200733. Moreover, we observed heterogeneity among the studies that analyze the association between rs2200733 and the risk of AF; this heterogeneity is partially explained by the study design.
Up to 9 genetic variants associated with the risk of AF have recently been identified.8 In our study, we analyzed only 2 of these variants (rs2200733 and rs7193343) because they are the ones most frequently identified in genome-wide association studies as being associated with AF and those in which this association is strongest.
Variant rs2200733 is found in the 4q25 region, where the PITX2 gene is also located. This gene is a transcription factor that plays a key role in the development of the fetal heart,27 and is also expressed in the adult heart, where it regulates the expression of genes related to ion channels.28PITX2 expression in atrial myocardial tissue has been found to be decreased in patients with AF, a circumstance that leads to atrial electrical and structural remodeling that favors arrhythmogenesis.29 Our findings in the Spanish population analyzed here agree closely with the results of the meta-analysis and indicate an association between the presence of the rare variant and greater probability of developing AF.
On the other hand, certain studies in murine models have found that carriers of the risk allele of polymorphism rs2200733 undergo atrial remodeling.29 In our study, the group of homozygous carriers of the risk allele of this variant had atrial enlargement, although this association disappeared when it was corrected for body surface area. We should point out that the available echocardiographic measurements of left atrium corresponded to a subgroup of patients and that the methodology utilized for measurement was not designed specifically to assess this association, as this was not among the objectives defined for this study. Properly designed studies are necessary to confirm the association between rs2200733 and atrial remodeling.
Variant rs7193343 is in the 16q22 region, where the ZFHX3 gene is located. This gene regulates neuronal and muscle differentiation and is a tumor suppressor gene in several types of cancer. Although it is expressed in mouse heart,30 its function in heart tissue is unknown. This variant is associated with AF in the meta-analysis, but not significantly in this Spanish population, probably because of the small size of the sample in our study and to the fact that the association observed is not as strong. With the sample size of our study (257 case patients and 379 controls), we were only able to identify as statistically significant those associations with an OR ≥ 1.63. There is another variant in the region, rs2106261, which is in linkage disequilibrium with rs7193343 (R2 = 0.77 in a Caucasian population from the HapMap-based study). It has also been analyzed in some studies and in 2 meta-analyses,7,8 and has a strength of association similar to that observed in our study (OR = 1.24 and OR = 1.22, respectively). Thus, the data indicate that both variants provide similar information.
Our study differs with respect to 2 characteristics. First, we analyzed the lone AF phenotype independently, observing that, in the case of rs2200733, the association is stronger than that observed for all types of AF.
Second, we analyzed the presence of heterogeneity across studies and its causes. We observed that the study design explains part of the heterogeneity observed in the association between rs2200733 and AF: the case-control studies have a stronger association than the cohort studies and, moreover, heterogeneity continues to be found between the case patients and controls. The heterogeneity across case-control studies is probably related to the varying quality of those studies,31 and the strength of the association observed in the case-control studies, which is probably greater than that of the cohort studies, is also a known phenomenon.31
Knowledge of the genetic bases of AF may contribute to the identification of new therapeutic targets. From the clinical point of view, it may also contribute to an improvement in the ability to predict the onset of AF in the future. A recent study performed with data from the Women's Health Study found that the inclusion of genetic information in the prediction model improves the discrimination and reclassification of AF risk.32 Another clinical application that is still pending examination and validation is the ability to identify those patients who will have a recurrence of AF following radiofrequency ablation.
LimitationsAmong the limitations of our study, we must point out that the size of the Spanish sample population is small, although the results agree with those of the meta-analysis performed. The distribution of ages and sexes in the control population differed from that of the case patients; this difference could influence the results. In any case, in the adjusted analyses and age- and sex-matched analyses, the results observed were very similar and consistent. The association between the genetic variants of interest and atrial remodeling has the limitations mentioned above, and it is necessary to confirm the findings in studies specifically designed to evaluate this hypothesis.
CONCLUSIONSGenetic variants rs2200733 and rs7193343 are associated with a higher risk of AF. These variants could have a clinical utility in estimating AF risk. On the other hand, case-control studies tend to overestimate the strength of the association between these genetic variants and the presence of AF.
FUNDINGThis study was financed by a grant for clinical research from the Spanish Society of Cardiology, Instituto de Salud Carlos III – European Regional Development Fund (Cardiovascular Research Network RD12/0042; PI09/90506), AGAUR (2009 SGR 1195).
CONFLICTS OF INTERESTNone declared.
The authors thank the Spanish National DNA Bank.